Abstract
Transfer functions of the conjugative plasmid RP4 (IncP alpha) are distributed among distinct regions of the genome, designated Tra1 and Tra2. By deletion analyses, we determined the limits of the Tra1 region, essential for intraspecific Escherichia coli matings. The Tra1 core region encompasses approximately 5.8 kb, including the genes traF, -G, -H, -I, -J, and -K as well as the origin of transfer. The traM gene product, however, is not absolutely required for conjugation but significantly increases transfer efficiency. To determine the transfer phenotype of genes encoded by the Tra2 core region, we generated a series of defined Tra2 mutants. This revealed that at least trbB, -C, -E, -G, and -L are essential for RP4 conjugation. To classify these transfer functions as components of the DNA transfer and replication (Dtr) or of the mating pair formation (Mpf) system, we analyzed the corresponding derivatives with respect to mobilization of IncQ plasmids and donor-specific phage propagation. We found that all of the Tra2 genes listed above and the traG and traF genes of Tra1 are required for RSF1010 mobilization. Expression of traF from Tra1 in conjunction with the Tra2 core was sufficient for phage propagation. This implies that the TraG protein is not directly involved in pilus formation and potentially connects the relaxosome with proteins enabling the membrane passage of the DNA. The proposed roles of the RP4 transfer gene products are discussed in the context of virulence functions encoded by the evolutionarily related Ti T-DNA transfer system of agrobacteria.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ayres E. K., Saadi S., Schreiner H. C., Thomson V. J., Figurski D. H. Differentiation of lethal and nonlethal, kor-regulated functions in the kilB region of broad host-range plasmid RK2. Plasmid. 1991 Jan;25(1):53–63. doi: 10.1016/0147-619x(91)90006-i. [DOI] [PubMed] [Google Scholar]
- Balzer D., Ziegelin G., Pansegrau W., Kruft V., Lanka E. KorB protein of promiscuous plasmid RP4 recognizes inverted sequence repetitions in regions essential for conjugative plasmid transfer. Nucleic Acids Res. 1992 Apr 25;20(8):1851–1858. doi: 10.1093/nar/20.8.1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barth P. T., Grinter N. J., Bradley D. E. Conjugal transfer system of plasmid RP4: analysis by transposon 7 insertion. J Bacteriol. 1978 Jan;133(1):43–52. doi: 10.1128/jb.133.1.43-52.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechhofer D. H., Kornacki J. A., Firshein W., Figurski D. H. Gene control in broad host range plasmid RK2: expression, polypeptide product, and multiple regulatory functions of korB. Proc Natl Acad Sci U S A. 1986 Jan;83(2):394–398. doi: 10.1073/pnas.83.2.394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beijersbergen A., Dulk-Ras A. D., Schilperoort R. A., Hooykaas P. J. Conjugative Transfer by the Virulence System of Agrobacterium tumefaciens. Science. 1992 May 29;256(5061):1324–1327. doi: 10.1126/science.256.5061.1324. [DOI] [PubMed] [Google Scholar]
- Bolland S., Llosa M., Avila P., de la Cruz F. General organization of the conjugal transfer genes of the IncW plasmid R388 and interactions between R388 and IncN and IncP plasmids. J Bacteriol. 1990 Oct;172(10):5795–5802. doi: 10.1128/jb.172.10.5795-5802.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer H. W., Roulland-Dussoix D. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J Mol Biol. 1969 May 14;41(3):459–472. doi: 10.1016/0022-2836(69)90288-5. [DOI] [PubMed] [Google Scholar]
- Cole S. P., Lanka E., Guiney D. G. Site-directed mutations in the relaxase operon of RP4. J Bacteriol. 1993 Aug;175(15):4911–4916. doi: 10.1128/jb.175.15.4911-4916.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engström P., Zambryski P., Van Montagu M., Stachel S. Characterization of Agrobacterium tumefaciens virulence proteins induced by the plant factor acetosyringone. J Mol Biol. 1987 Oct 20;197(4):635–645. doi: 10.1016/0022-2836(87)90470-0. [DOI] [PubMed] [Google Scholar]
- Figurski D. H., Pohlman R. F., Bechhofer D. H., Prince A. S., Kelton C. A. Broad host range plasmid RK2 encodes multiple kil genes potentially lethal to Escherichia coli host cells. Proc Natl Acad Sci U S A. 1982 Mar;79(6):1935–1939. doi: 10.1073/pnas.79.6.1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figurski D., Meyer R., Miller D. S., Helinski D. R. Generation in vitro of deletions in the broad host range plasmid RK2 using phage Mu insertions and a restriction endonuclease. Gene. 1976;1(1):107–119. doi: 10.1016/0378-1119(76)90010-x. [DOI] [PubMed] [Google Scholar]
- Fong S. T., Stanisich V. A. Identification and characterization of RP1 Tra1 cistrons involved in pilus function and plasmid mobilization. J Bacteriol. 1993 Jan;175(2):448–456. doi: 10.1128/jb.175.2.448-456.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fürste J. P., Pansegrau W., Frank R., Blöcker H., Scholz P., Bagdasarian M., Lanka E. Molecular cloning of the plasmid RP4 primase region in a multi-host-range tacP expression vector. Gene. 1986;48(1):119–131. doi: 10.1016/0378-1119(86)90358-6. [DOI] [PubMed] [Google Scholar]
- Fürste J. P., Pansegrau W., Ziegelin G., Kröger M., Lanka E. Conjugative transfer of promiscuous IncP plasmids: interaction of plasmid-encoded products with the transfer origin. Proc Natl Acad Sci U S A. 1989 Mar;86(6):1771–1775. doi: 10.1073/pnas.86.6.1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goncharoff P., Saadi S., Chang C. H., Saltman L. H., Figurski D. H. Structural, molecular, and genetic analysis of the kilA operon of broad-host-range plasmid RK2. J Bacteriol. 1991 Jun;173(11):3463–3477. doi: 10.1128/jb.173.11.3463-3477.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guiney D. G., Deiss C., Simnad V., Yee L., Pansegrau W., Lanka E. Mutagenesis of the Tra1 core region of RK2 by using Tn5: identification of plasmid-specific transfer genes. J Bacteriol. 1989 Jul;171(7):4100–4103. doi: 10.1128/jb.171.7.4100-4103.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983 Jun 5;166(4):557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- Heinemann J. A., Sprague G. F., Jr Bacterial conjugative plasmids mobilize DNA transfer between bacteria and yeast. Nature. 1989 Jul 20;340(6230):205–209. doi: 10.1038/340205a0. [DOI] [PubMed] [Google Scholar]
- Ippen-Ihler K. A., Minkley E. G., Jr The conjugation system of F, the fertility factor of Escherichia coli. Annu Rev Genet. 1986;20:593–624. doi: 10.1146/annurev.ge.20.120186.003113. [DOI] [PubMed] [Google Scholar]
- Jagura-Burdzy G., Ibbotson J. P., Thomas C. M. The korF region of broad-host-range plasmid RK2 encodes two polypeptides with transcriptional repressor activity. J Bacteriol. 1991 Jan;173(2):826–833. doi: 10.1128/jb.173.2.826-833.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagura-Burdzy G., Khanim F., Smith C. A., Thomas C. M. Crosstalk between plasmid vegetative replication and conjugative transfer: repression of the trfA operon by trbA of broad host range plasmid RK2. Nucleic Acids Res. 1992 Aug 11;20(15):3939–3944. doi: 10.1093/nar/20.15.3939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jovanovic O. S., Ayres E. K., Figurski D. H. The replication initiator operon of promiscuous plasmid RK2 encodes a gene that complements an Escherichia coli mutant defective in single-stranded DNA-binding protein. J Bacteriol. 1992 Jul;174(14):4842–4846. doi: 10.1128/jb.174.14.4842-4846.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornacki J. A., Balderes P. J., Figurski D. H. Nucleotide sequence of korB, a replication control gene of broad host-range plasmid RK2. J Mol Biol. 1987 Nov 20;198(2):211–222. doi: 10.1016/0022-2836(87)90307-x. [DOI] [PubMed] [Google Scholar]
- Kotilainen M. M., Grahn A. M., Bamford J. K., Bamford D. H. Binding of an Escherichia coli double-stranded DNA virus PRD1 to a receptor coded by an IncP-type plasmid. J Bacteriol. 1993 May;175(10):3089–3095. doi: 10.1128/jb.175.10.3089-3095.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanka E., Barth P. T. Plasmid RP4 specifies a deoxyribonucleic acid primase involved in its conjugal transfer and maintenance. J Bacteriol. 1981 Dec;148(3):769–781. doi: 10.1128/jb.148.3.769-781.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanka E., Lurz R., Fürste J. P. Molecular cloning and mapping of SphI restriction fragments of plasmid RP4. Plasmid. 1983 Nov;10(3):303–307. doi: 10.1016/0147-619x(83)90047-1. [DOI] [PubMed] [Google Scholar]
- Lanka E., Lurz R., Kröger M., Fürste J. P. Plasmid RP4 encodes two forms of a DNA primase. Mol Gen Genet. 1984;194(1-2):65–72. doi: 10.1007/BF00383499. [DOI] [PubMed] [Google Scholar]
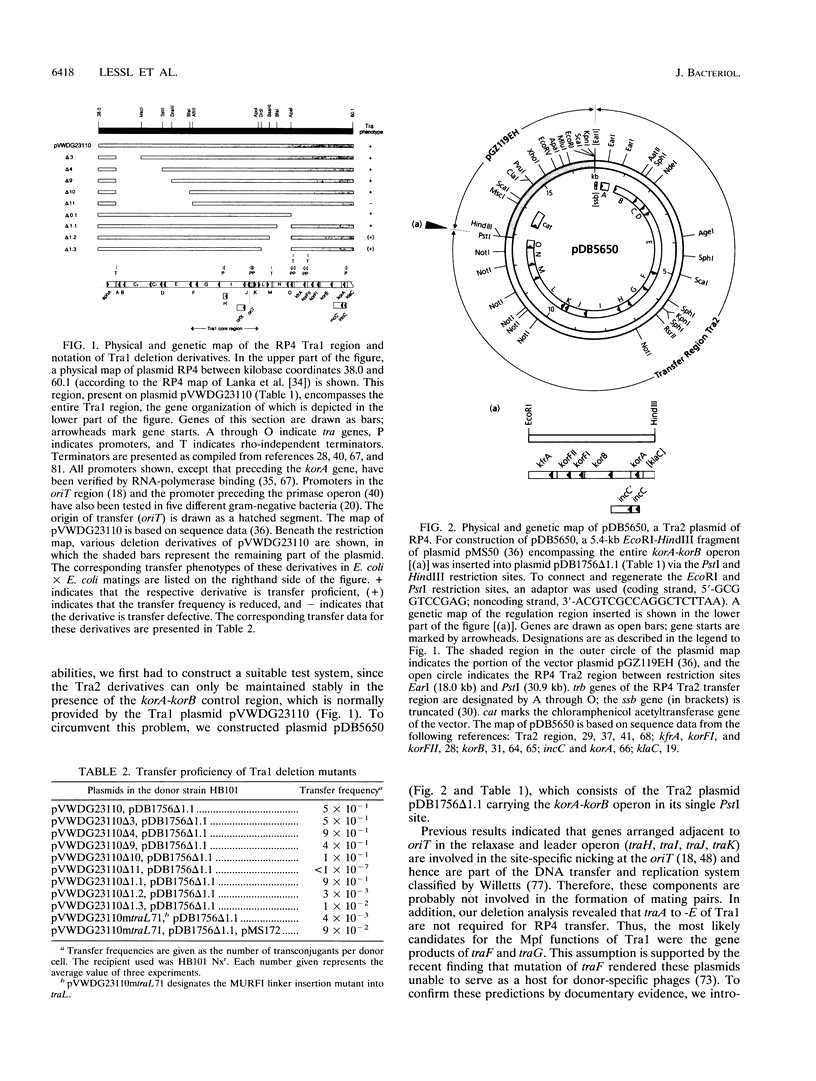
- Lessl M., Balzer D., Lurz R., Waters V. L., Guiney D. G., Lanka E. Dissection of IncP conjugative plasmid transfer: definition of the transfer region Tra2 by mobilization of the Tra1 region in trans. J Bacteriol. 1992 Apr;174(8):2493–2500. doi: 10.1128/jb.174.8.2493-2500.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lessl M., Balzer D., Pansegrau W., Lanka E. Sequence similarities between the RP4 Tra2 and the Ti VirB region strongly support the conjugation model for T-DNA transfer. J Biol Chem. 1992 Oct 5;267(28):20471–20480. [PubMed] [Google Scholar]
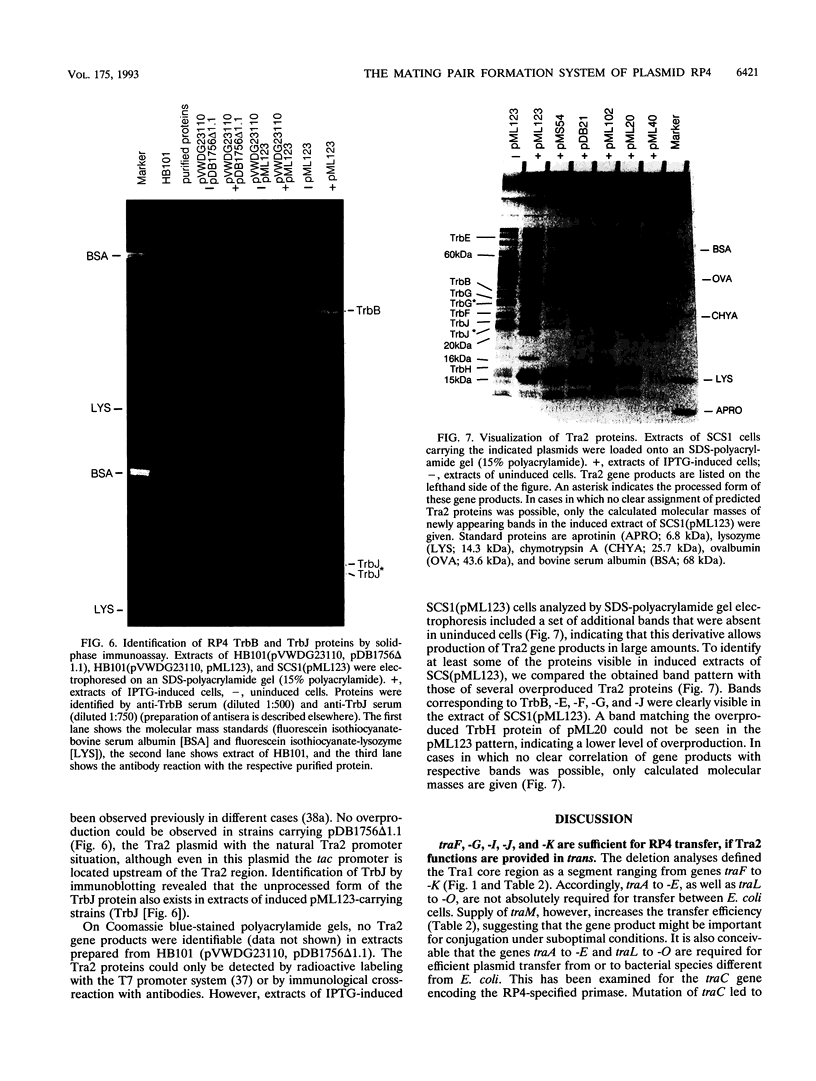
- Lessl M., Krishnapillai V., Schilf W. Identification and characterization of two entry exclusion genes of the promiscuous IncP plasmid R18. Mol Gen Genet. 1991 May;227(1):120–126. doi: 10.1007/BF00260716. [DOI] [PubMed] [Google Scholar]
- Lessl M., Pansegrau W., Lanka E. Relationship of DNA-transfer-systems: essential transfer factors of plasmids RP4, Ti and F share common sequences. Nucleic Acids Res. 1992 Nov 25;20(22):6099–6100. doi: 10.1093/nar/20.22.6099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miele L., Strack B., Kruft V., Lanka E. Gene organization and nucleotide sequence of the primase region of IncP plasmids RP4 and R751. DNA Seq. 1991;2(3):145–162. doi: 10.3109/10425179109039685. [DOI] [PubMed] [Google Scholar]
- Motallebi-Veshareh M., Balzer D., Lanka E., Jagura-Burdzy G., Thomas C. M. Conjugative transfer functions of broad-host-range plasmid RK2 are coregulated with vegetative replication. Mol Microbiol. 1992 Apr;6(7):907–920. doi: 10.1111/j.1365-2958.1992.tb01541.x. [DOI] [PubMed] [Google Scholar]
- Oka A., Sugisaki H., Takanami M. Nucleotide sequence of the kanamycin resistance transposon Tn903. J Mol Biol. 1981 Apr 5;147(2):217–226. doi: 10.1016/0022-2836(81)90438-1. [DOI] [PubMed] [Google Scholar]
- Okamoto S., Toyoda-Yamamoto A., Ito K., Takebe I., Machida Y. Localization and orientation of the VirD4 protein of Agrobacterium tumefaciens in the cell membrane. Mol Gen Genet. 1991 Aug;228(1-2):24–32. doi: 10.1007/BF00282443. [DOI] [PubMed] [Google Scholar]
- Olsen R. H., Shipley P. Host range and properties of the Pseudomonas aeruginosa R factor R1822. J Bacteriol. 1973 Feb;113(2):772–780. doi: 10.1128/jb.113.2.772-780.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen R. H., Siak J. S., Gray R. H. Characteristics of PRD1, a plasmid-dependent broad host range DNA bacteriophage. J Virol. 1974 Sep;14(3):689–699. doi: 10.1128/jvi.14.3.689-699.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen R. H., Thomas D. D. Characteristics and purification of PRR1, an RNA phage specific for the broad host range Pseudomonas R1822 drug resistance plasmid. J Virol. 1973 Dec;12(6):1560–1567. doi: 10.1128/jvi.12.6.1560-1567.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panicker M. M., Minkley E. G., Jr Purification and properties of the F sex factor TraD protein, an inner membrane conjugal transfer protein. J Biol Chem. 1992 Jun 25;267(18):12761–12766. [PubMed] [Google Scholar]
- Pansegrau W., Balzer D., Kruft V., Lurz R., Lanka E. In vitro assembly of relaxosomes at the transfer origin of plasmid RP4. Proc Natl Acad Sci U S A. 1990 Sep;87(17):6555–6559. doi: 10.1073/pnas.87.17.6555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pansegrau W., Lanka E. Common sequence motifs in DNA relaxases and nick regions from a variety of DNA transfer systems. Nucleic Acids Res. 1991 Jun 25;19(12):3455–3455. doi: 10.1093/nar/19.12.3455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pansegrau W., Schröder W., Lanka E. Relaxase (TraI) of IncP alpha plasmid RP4 catalyzes a site-specific cleaving-joining reaction of single-stranded DNA. Proc Natl Acad Sci U S A. 1993 Apr 1;90(7):2925–2929. doi: 10.1073/pnas.90.7.2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pansegrau W., Ziegelin G., Lanka E. Covalent association of the traI gene product of plasmid RP4 with the 5'-terminal nucleotide at the relaxation nick site. J Biol Chem. 1990 Jun 25;265(18):10637–10644. [PubMed] [Google Scholar]
- Perlman D., Halvorson H. O. The MURFI linker for multiple reading frame insertion of a sense or nonsense codon into DNA. Nucleic Acids Res. 1986 Mar 11;14(5):2139–2155. doi: 10.1093/nar/14.5.2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rees C. E., Wilkins B. M. Protein transfer into the recipient cell during bacterial conjugation: studies with F and RP4. Mol Microbiol. 1990 Jul;4(7):1199–1205. doi: 10.1111/j.1365-2958.1990.tb00695.x. [DOI] [PubMed] [Google Scholar]
- Rohrer J., Rawlings D. E. Sequence analysis and characterization of the mobilization region of a broad-host-range plasmid, pTF-FC2, isolated from Thiobacillus ferrooxidans. J Bacteriol. 1992 Oct;174(19):6230–6237. doi: 10.1128/jb.174.19.6230-6237.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholz P., Haring V., Wittmann-Liebold B., Ashman K., Bagdasarian M., Scherzinger E. Complete nucleotide sequence and gene organization of the broad-host-range plasmid RSF1010. Gene. 1989 Feb 20;75(2):271–288. doi: 10.1016/0378-1119(89)90273-4. [DOI] [PubMed] [Google Scholar]
- Shirasu K., Kado C. I. The virB operon of the Agrobacterium tumefaciens virulence regulon has sequence similarities to B, C and D open reading frames downstream of the pertussis toxin-operon and to the DNA transfer-operons of broad-host-range conjugative plasmids. Nucleic Acids Res. 1993 Jan 25;21(2):353–354. doi: 10.1093/nar/21.2.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith C. A., Shingler V., Thomas C. M. The trfA and trfB promoter regions of broad host range plasmid RK2 share common potential regulatory sequences. Nucleic Acids Res. 1984 Apr 25;12(8):3619–3630. doi: 10.1093/nar/12.8.3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith C. A., Thomas C. M. Nucleotide sequence of the trfA gene of broad host-range plasmid RK2. J Mol Biol. 1984 May 25;175(3):251–262. doi: 10.1016/0022-2836(84)90347-4. [DOI] [PubMed] [Google Scholar]
- Stanisich V. A. The properties and host range of male-specific bacteriophages of Pseudomonas aeruginosa. J Gen Microbiol. 1974 Oct;84(2):332–342. doi: 10.1099/00221287-84-2-332. [DOI] [PubMed] [Google Scholar]
- Stokes H. W., Moore R. J., Krishnapillai V. Complementation analysis in Pseudomonas aeruginosa of the transfer genes of the wide host range R plasmid R18. Plasmid. 1981 Mar;5(2):202–212. doi: 10.1016/0147-619x(81)90021-4. [DOI] [PubMed] [Google Scholar]
- Strack B., Lessl M., Calendar R., Lanka E. A common sequence motif, -E-G-Y-A-T-A-, identified within the primase domains of plasmid-encoded I- and P-type DNA primases and the alpha protein of the Escherichia coli satellite phage P4. J Biol Chem. 1992 Jun 25;267(18):13062–13072. [PubMed] [Google Scholar]
- Theophilus B. D., Thomas C. M. Nucleotide sequence of the transcriptional repressor gene korB which plays a key role in regulation of the copy number of broad host range plasmid RK2. Nucleic Acids Res. 1987 Sep 25;15(18):7443–7450. doi: 10.1093/nar/15.18.7443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas C. M., Smith C. A. The trfB region of broad host range plasmid RK2: the nucleotide sequence reveals incC and key regulatory gene trfB/korA/korD as overlapping genes. Nucleic Acids Res. 1986 Jun 11;14(11):4453–4469. doi: 10.1093/nar/14.11.4453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas C. M., Theophilus B. D., Johnston L., Jagura-Burdzy G., Schilf W., Lurz R., Lanka E. Identification of a seventh operon on plasmid RK2 regulated by the korA gene product. Gene. 1990 Apr 30;89(1):29–35. doi: 10.1016/0378-1119(90)90202-3. [DOI] [PubMed] [Google Scholar]
- Thomson V. J., Jovanovic O. S., Pohlman R. F., Chang C. H., Figurski D. H. Structure, function, and regulation of the kilB locus of promiscuous plasmid RK2. J Bacteriol. 1993 Apr;175(8):2423–2435. doi: 10.1128/jb.175.8.2423-2435.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towbin H., Staehelin T., Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward J. E., Akiyoshi D. E., Regier D., Datta A., Gordon M. P., Nester E. W. Characterization of the virB operon from an Agrobacterium tumefaciens Ti plasmid. J Biol Chem. 1988 Apr 25;263(12):5804–5814. [PubMed] [Google Scholar]
- Ward J. E., Akiyoshi D. E., Regier D., Datta A., Gordon M. P., Nester E. W. Correction: characterization of the virB operon from Agrobacterium tumefaciens Ti plasmid. J Biol Chem. 1990 Mar 15;265(8):4768–4768. [PubMed] [Google Scholar]
- Waters V. L., Hirata K. H., Pansegrau W., Lanka E., Guiney D. G. Sequence identity in the nick regions of IncP plasmid transfer origins and T-DNA borders of Agrobacterium Ti plasmids. Proc Natl Acad Sci U S A. 1991 Feb 15;88(4):1456–1460. doi: 10.1073/pnas.88.4.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters V. L., Strack B., Pansegrau W., Lanka E., Guiney D. G. Mutational analysis of essential IncP alpha plasmid transfer genes traF and traG and involvement of traF in phage sensitivity. J Bacteriol. 1992 Oct;174(20):6666–6673. doi: 10.1128/jb.174.20.6666-6673.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson J., Schmidt L., Willetts N. Cloning the Tra1 region of RP1. Plasmid. 1980 Sep;4(2):175–183. doi: 10.1016/0147-619x(80)90007-4. [DOI] [PubMed] [Google Scholar]
- Weiss A. A., Johnson F. D., Burns D. L. Molecular characterization of an operon required for pertussis toxin secretion. Proc Natl Acad Sci U S A. 1993 Apr 1;90(7):2970–2974. doi: 10.1073/pnas.90.7.2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willetts N., Crowther C. Mobilization of the non-conjugative IncQ plasmid RSF1010. Genet Res. 1981 Jun;37(3):311–316. doi: 10.1017/s0016672300020310. [DOI] [PubMed] [Google Scholar]
- Ziegelin G., Fürste J. P., Lanka E. TraJ protein of plasmid RP4 binds to a 19-base pair invert sequence repetition within the transfer origin. J Biol Chem. 1989 Jul 15;264(20):11989–11994. [PubMed] [Google Scholar]
- Ziegelin G., Pansegrau W., Lurz R., Lanka E. TraK protein of conjugative plasmid RP4 forms a specialized nucleoprotein complex with the transfer origin. J Biol Chem. 1992 Aug 25;267(24):17279–17286. [PubMed] [Google Scholar]
- Ziegelin G., Pansegrau W., Strack B., Balzer D., Kröger M., Kruft V., Lanka E. Nucleotide sequence and organization of genes flanking the transfer origin of promiscuous plasmid RP4. DNA Seq. 1991;1(5):303–327. doi: 10.3109/10425179109020786. [DOI] [PubMed] [Google Scholar]







