Abstract
Most Bacillus subtilis tRNA genes have been isolated from lambda libraries by use of probes that hybridize to tRNA or rRNA sequences. None of those genes map to the region of the sup-3 mutation. By cloning of the sup-3 allele, a cluster of seven tRNA genes (the trnS operon) that had not been isolated by other methods was identified. In principle, this approach could be used to isolate at least one more predicted tRNA-containing operon in this bacterium. The trnS operon was shown to contain tRNA genes for Asn (GUU), Ser (GCU), Glu (UUC), Gln (UUG), Lys (UUU), Leu (UAG), and Leu (GAG). The sup-3 mutation was found to be a T-to-A transversion that changes the anticodon of the lysine tRNA from 5'-UUU-3' to 5'-UUA-3'. This result agrees with previous work that determined that the sup-3 mutation causes lysine to be inserted at ochre nonsense mutations.
Full text
PDF
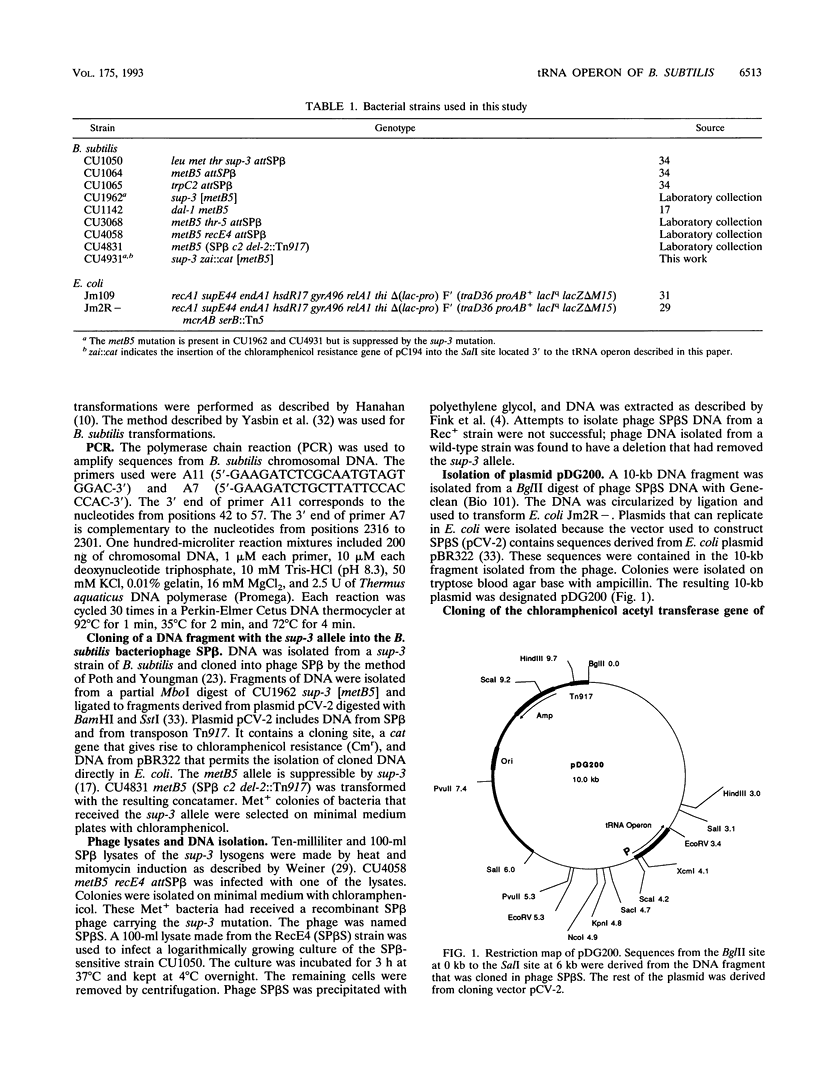
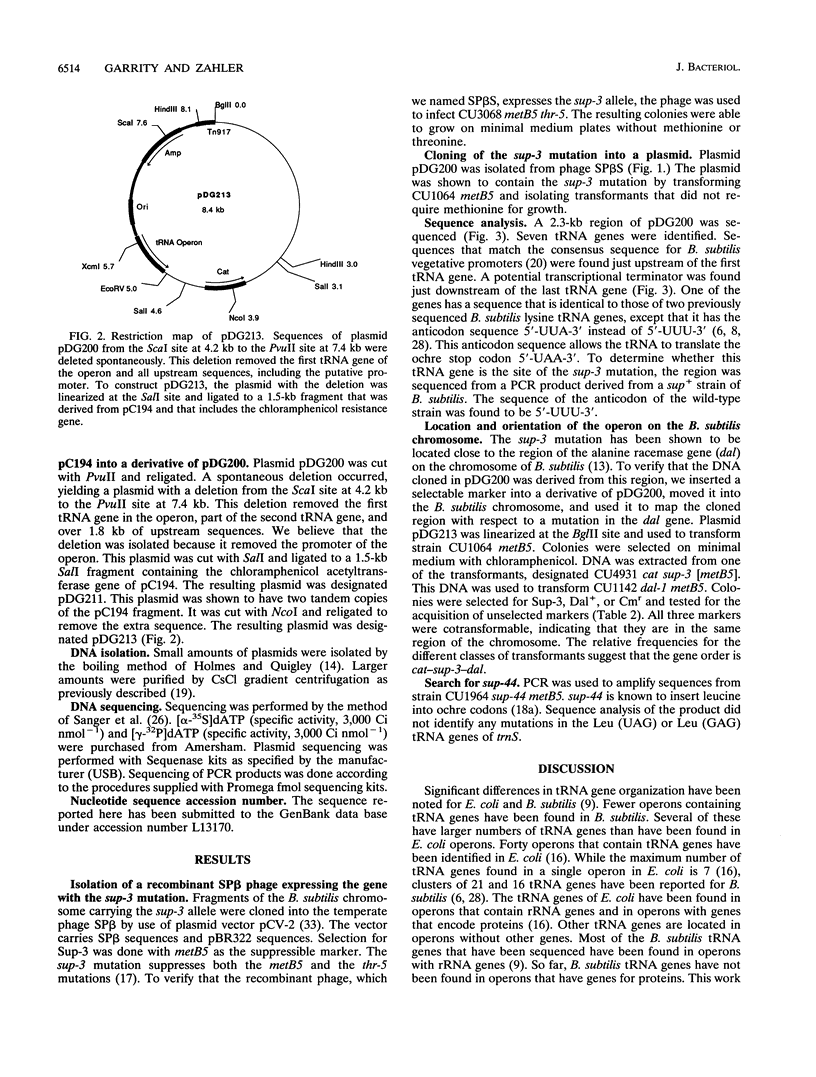



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barrell B. G., Anderson S., Bankier A. T., de Bruijn M. H., Chen E., Coulson A. R., Drouin J., Eperon I. C., Nierlich D. P., Roe B. A. Different pattern of codon recognition by mammalian mitochondrial tRNAs. Proc Natl Acad Sci U S A. 1980 Jun;77(6):3164–3166. doi: 10.1073/pnas.77.6.3164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonitz S. G., Berlani R., Coruzzi G., Li M., Macino G., Nobrega F. G., Nobrega M. P., Thalenfeld B. E., Tzagoloff A. Codon recognition rules in yeast mitochondria. Proc Natl Acad Sci U S A. 1980 Jun;77(6):3167–3170. doi: 10.1073/pnas.77.6.3167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fink P. S., Korman R. Z., Odebralski J. M., Zahler S. A. Bacillus subtilis bacteriophages SP beta c1 is a deletion mutant of SP beta. Mol Gen Genet. 1981;182(3):514–515. doi: 10.1007/BF00293946. [DOI] [PubMed] [Google Scholar]
- Georgopoulos C. P. Suppressor system in Bacillus subtilis 168. J Bacteriol. 1969 Mar;97(3):1397–1402. doi: 10.1128/jb.97.3.1397-1402.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green C. J., Stewart G. C., Hollis M. A., Vold B. S., Bott K. F. Nucleotide sequence of the Bacillus subtilis ribosomal RNA operon, rrnB. Gene. 1985;37(1-3):261–266. doi: 10.1016/0378-1119(85)90281-1. [DOI] [PubMed] [Google Scholar]
- Green C. J., Vold B. S. A cluster of nine tRNA genes between ribosomal gene operons in Bacillus subtilis. J Bacteriol. 1992 May;174(10):3147–3151. doi: 10.1128/jb.174.10.3147-3151.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green C. J., Vold B. S. Sequence analysis of a cluster of twenty-one tRNA genes in Bacillus subtilis. Nucleic Acids Res. 1983 Aug 25;11(16):5763–5774. doi: 10.1093/nar/11.16.5763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983 Jun 5;166(4):557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- Heckman J. E., Sarnoff J., Alzner-DeWeerd B., Yin S., RajBhandary U. L. Novel features in the genetic code and codon reading patterns in Neurospora crassa mitochondria based on sequences of six mitochondrial tRNAs. Proc Natl Acad Sci U S A. 1980 Jun;77(6):3159–3163. doi: 10.1073/pnas.77.6.3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henner D. J., Steinberg W. Genetic location of the Bacillus subtilis sup-3 suppressor mutation. J Bacteriol. 1979 Aug;139(2):668–670. doi: 10.1128/jb.139.2.668-670.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes D. S., Quigley M. A rapid boiling method for the preparation of bacterial plasmids. Anal Biochem. 1981 Jun;114(1):193–197. doi: 10.1016/0003-2697(81)90473-5. [DOI] [PubMed] [Google Scholar]
- Komine Y., Adachi T., Inokuchi H., Ozeki H. Genomic organization and physical mapping of the transfer RNA genes in Escherichia coli K12. J Mol Biol. 1990 Apr 20;212(4):579–598. doi: 10.1016/0022-2836(90)90224-A. [DOI] [PubMed] [Google Scholar]
- Lipsky R. H., Rosenthal R., Zahler S. A. Defective specialized SP beta transducing bacteriophages of Bacillus subtilis that carry the sup-3 or sup-44 gene. J Bacteriol. 1981 Dec;148(3):1012–1015. doi: 10.1128/jb.148.3.1012-1015.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loughney K., Lund E., Dahlberg J. E. tRNA genes are found between 16S and 23S rRNA genes in Bacillus subtilis. Nucleic Acids Res. 1982 Mar 11;10(5):1607–1624. doi: 10.1093/nar/10.5.1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovett P. S., Ambulos N. P., Jr, Mulbry W., Noguchi N., Rogers E. J. UGA can be decoded as tryptophan at low efficiency in Bacillus subtilis. J Bacteriol. 1991 Mar;173(5):1810–1812. doi: 10.1128/jb.173.5.1810-1812.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran C. P., Jr, Lang N., LeGrice S. F., Lee G., Stephens M., Sonenshein A. L., Pero J., Losick R. Nucleotide sequences that signal the initiation of transcription and translation in Bacillus subtilis. Mol Gen Genet. 1982;186(3):339–346. doi: 10.1007/BF00729452. [DOI] [PubMed] [Google Scholar]
- Mulbry W. W., Ambulos N. P., Jr, Lovett P. S. Bacillus subtilis mutant allele sup-3 causes lysine insertion at ochre codons: use of sup-3 in studies of translational attenuation. J Bacteriol. 1989 Oct;171(10):5322–5324. doi: 10.1128/jb.171.10.5322-5324.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogasawara N., Moriya S., Yoshikawa H. Structure and organization of rRNA operons in the region of the replication origin of the Bacillus subtilis chromosome. Nucleic Acids Res. 1983 Sep 24;11(18):6301–6318. doi: 10.1093/nar/11.18.6301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poth H., Youngman P. A new cloning system for Bacillus subtilis comprising elements of phage, plasmid and transposon vectors. Gene. 1988 Dec 15;73(1):215–226. doi: 10.1016/0378-1119(88)90328-9. [DOI] [PubMed] [Google Scholar]
- Rudner R., Chevrestt A., Buchholz S. R., Studamire B., White A. M., Jarvis E. D. Two tRNA gene clusters associated with rRNA operons rrnD and rrnE in Bacillus subtilis. J Bacteriol. 1993 Jan;175(2):503–509. doi: 10.1128/jb.175.2.503-509.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuelsson T., Guindy Y. S., Lustig F., Borén T., Lagerkvist U. Apparent lack of discrimination in the reading of certain codons in Mycoplasma mycoides. Proc Natl Acad Sci U S A. 1987 May;84(10):3166–3170. doi: 10.1073/pnas.84.10.3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wawrousek E. F., Hansen J. N. Structure and organization of a cluster of sic tRNA genes in the space between tandem ribosomal RNA gene sets in Bacillus subtilis. J Biol Chem. 1983 Jan 10;258(1):291–298. [PubMed] [Google Scholar]
- Wawrousek E. F., Narasimhan N., Hansen J. N. Two large clusters with thirty-seven transfer RNA genes adjacent to ribosomal RNA gene sets in Bacillus subtilis. Sequence and organization of trrnD and trrnE gene clusters. J Biol Chem. 1984 Mar 25;259(6):3694–3702. [PubMed] [Google Scholar]
- Yamada Y., Ohki M., Ishikura H. The nucleotide sequence of Bacillus subtilis tRNA genes. Nucleic Acids Res. 1983 May 25;11(10):3037–3045. doi: 10.1093/nar/11.10.3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- Yasbin R. E., Wilson G. A., Young F. E. Transformation and transfection in lysogenic strains of Bacillus subtilis: evidence for selective induction of prophage in competent cells. J Bacteriol. 1975 Jan;121(1):296–304. doi: 10.1128/jb.121.1.296-304.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahler S. A., Korman R. Z., Rosenthal R., Hemphill H. E. Bacillus subtilis bacteriophage SPbeta: localization of the prophage attachment site, and specialized transduction. J Bacteriol. 1977 Jan;129(1):556–558. doi: 10.1128/jb.129.1.556-558.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]


