Abstract
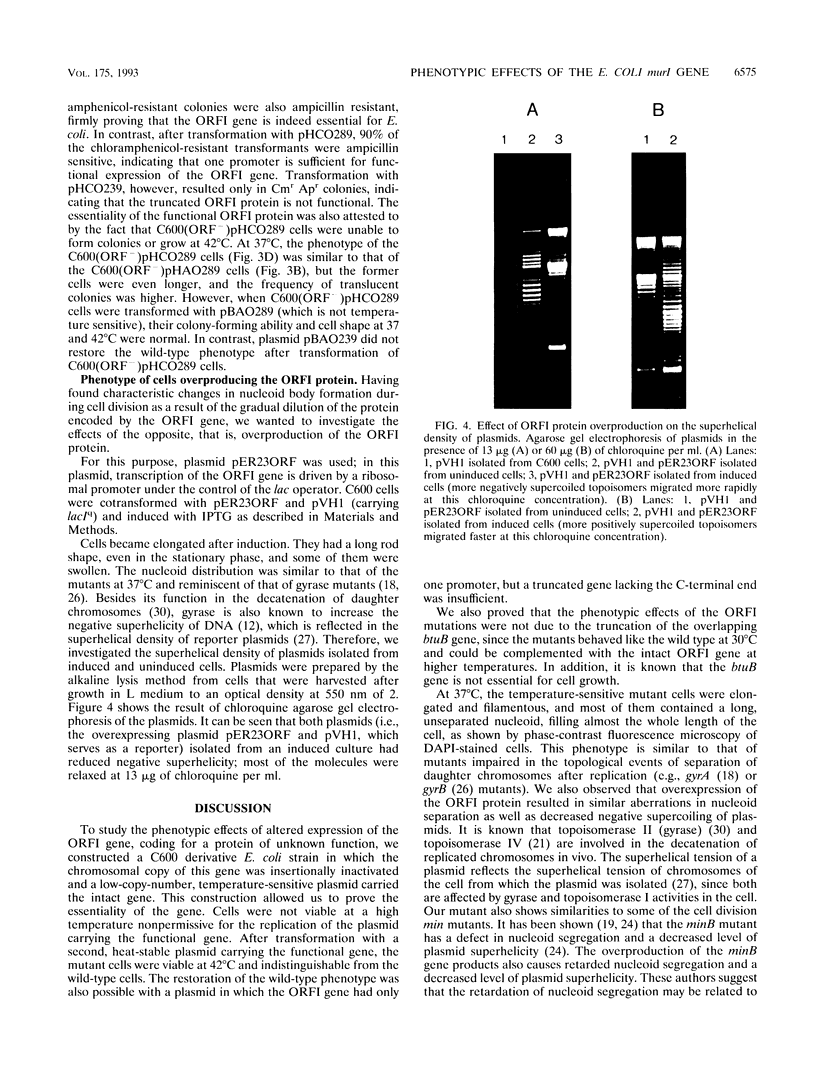
Earlier we reported that an open reading frame located at 89.5 min of the Escherichia coli map (ORFI) codes for a protein of unknown function that could be overexpressed and purified to homogeneity (G. Balikó, A. Raukas, I. Boros, and P. Venetianer, Mol. Gen. Genet. 211:326-331, 1988). In the work described here, we attempted to learn the function of this protein by inactivating the chromosomal gene and providing it or its deletion derivatives on temperature-sensitive plasmids. We found that the presence of the functional ORFI gene is essential; cells are not viable at the nonpermissive temperature or when the region coding for the C-terminal 50 amino acids of the protein is deleted. At intermediate temperatures or when the gene is overexpressed, characteristic changes occur in cell morphology, nucleoid separation during cell division, and supercoiling of plasmids. The possible mechanisms of these effects are discussed in view of the fact that Doublet et al. (P. Doublet, J. van Heijenoort, and D. Mengin-Lecreulx, J. Bacteriol. 174:5772-5779, 1992) recently identified the ORFI gene as murI, involved in D-glutamic acid biosynthesis.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Appleyard R K. Segregation of New Lysogenic Types during Growth of a Doubly Lysogenic Strain Derived from Escherichia Coli K12. Genetics. 1954 Jul;39(4):440–452. doi: 10.1093/genetics/39.4.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balikó G., Raukas A., Boros I., Venetianer P. An Escherichia coli gene in search of a function. Mol Gen Genet. 1988 Feb;211(2):326–331. doi: 10.1007/BF00330611. [DOI] [PubMed] [Google Scholar]
- Boros I., Csordás-Tóth E., Kiss A., Kiss I., Török I., Udvardy A., Udvardy K., Venetianer P. Identification of two new promoters probably involved in the transcription of a ribosomal RNA gene of Escherichia coli. Biochim Biophys Acta. 1983 Mar 10;739(2):173–180. doi: 10.1016/0167-4781(83)90027-1. [DOI] [PubMed] [Google Scholar]
- Boros I., Kiss A., Sain B., Somlyai G., Venetianer P. Cloning of the promoters of an Escherichia coli rRNA gene. New experimental system to study the regulation of rRNA transcription. Gene. 1983 May-Jun;22(2-3):191–201. doi: 10.1016/0378-1119(83)90103-8. [DOI] [PubMed] [Google Scholar]
- Boros I., Pósfai G., Venetianer P. High-copy-number derivatives of the plasmid cloning vector pBR322. Gene. 1984 Oct;30(1-3):257–260. doi: 10.1016/0378-1119(84)90130-6. [DOI] [PubMed] [Google Scholar]
- Boyer H. W., Roulland-Dussoix D. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J Mol Biol. 1969 May 14;41(3):459–472. doi: 10.1016/0022-2836(69)90288-5. [DOI] [PubMed] [Google Scholar]
- Brosius J., Dull T. J., Sleeter D. D., Noller H. F. Gene organization and primary structure of a ribosomal RNA operon from Escherichia coli. J Mol Biol. 1981 May 15;148(2):107–127. doi: 10.1016/0022-2836(81)90508-8. [DOI] [PubMed] [Google Scholar]
- Doublet P., van Heijenoort J., Bohin J. P., Mengin-Lecreulx D. The murI gene of Escherichia coli is an essential gene that encodes a glutamate racemase activity. J Bacteriol. 1993 May;175(10):2970–2979. doi: 10.1128/jb.175.10.2970-2979.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doublet P., van Heijenoort J., Mengin-Lecreulx D. Identification of the Escherichia coli murI gene, which is required for the biosynthesis of D-glutamic acid, a specific component of bacterial peptidoglycan. J Bacteriol. 1992 Sep;174(18):5772–5779. doi: 10.1128/jb.174.18.5772-5779.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dougherty T. J., Thanassi J. A., Pucci M. J. The Escherichia coli mutant requiring D-glutamic acid is the result of mutations in two distinct genetic loci. J Bacteriol. 1993 Jan;175(1):111–116. doi: 10.1128/jb.175.1.111-116.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983 Jul 1;132(1):6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Gellert M., Mizuuchi K., O'Dea M. H., Nash H. A. DNA gyrase: an enzyme that introduces superhelical turns into DNA. Proc Natl Acad Sci U S A. 1976 Nov;73(11):3872–3876. doi: 10.1073/pnas.73.11.3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton C. M., Aldea M., Washburn B. K., Babitzke P., Kushner S. R. New method for generating deletions and gene replacements in Escherichia coli. J Bacteriol. 1989 Sep;171(9):4617–4622. doi: 10.1128/jb.171.9.4617-4622.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haring V., Scholz P., Scherzinger E., Frey J., Derbyshire K., Hatfull G., Willetts N. S., Bagdasarian M. Protein RepC is involved in copy number control of the broad host range plasmid RSF1010. Proc Natl Acad Sci U S A. 1985 Sep;82(18):6090–6094. doi: 10.1073/pnas.82.18.6090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto-Gotoh T., Franklin F. C., Nordheim A., Timmis K. N. Specific-purpose plasmid cloning vectors. I. Low copy number, temperature-sensitive, mobilization-defective pSC101-derived containment vectors. Gene. 1981 Dec;16(1-3):227–235. doi: 10.1016/0378-1119(81)90079-2. [DOI] [PubMed] [Google Scholar]
- Hiraga S., Niki H., Ogura T., Ichinose C., Mori H., Ezaki B., Jaffé A. Chromosome partitioning in Escherichia coli: novel mutants producing anucleate cells. J Bacteriol. 1989 Mar;171(3):1496–1505. doi: 10.1128/jb.171.3.1496-1505.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann B., Messer W., Schwarz U. Regulation of polar cap formation in the life cycle of Escherichia coli. J Supramol Struct. 1972;1(1):29–37. doi: 10.1002/jss.400010105. [DOI] [PubMed] [Google Scholar]
- Hussain K., Begg K. J., Salmond G. P., Donachie W. D. ParD: a new gene coding for a protein required for chromosome partitioning and septum localization in Escherichia coli. Mol Microbiol. 1987 Jul;1(1):73–81. doi: 10.1111/j.1365-2958.1987.tb00529.x. [DOI] [PubMed] [Google Scholar]
- Jaffé A., D'Ari R., Hiraga S. Minicell-forming mutants of Escherichia coli: production of minicells and anucleate rods. J Bacteriol. 1988 Jul;170(7):3094–3101. doi: 10.1128/jb.170.7.3094-3101.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalman M., Gentry D. R., Cashel M. Characterization of the Escherichia coli K12 gltS glutamate permease gene. Mol Gen Genet. 1991 Mar;225(3):379–386. doi: 10.1007/BF00261677. [DOI] [PubMed] [Google Scholar]
- Kato J., Nishimura Y., Imamura R., Niki H., Hiraga S., Suzuki H. New topoisomerase essential for chromosome segregation in E. coli. Cell. 1990 Oct 19;63(2):393–404. doi: 10.1016/0092-8674(90)90172-b. [DOI] [PubMed] [Google Scholar]
- Lugtenberg E. J., Wijsman H. J., van Zaane D. Properties of a D-glutamic acid-requiring mutant of Escherichia coli. J Bacteriol. 1973 May;114(2):499–506. doi: 10.1128/jb.114.2.499-506.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulder E., El'Bouhali M., Pas E., Woldringh C. L. The Escherichia coli minB mutation resembles gyrB in defective nucleoid segregation and decreased negative supercoiling of plasmids. Mol Gen Genet. 1990 Mar;221(1):87–93. doi: 10.1007/BF00280372. [DOI] [PubMed] [Google Scholar]
- Mulder E., Woldringh C. L. Actively replicating nucleoids influence positioning of division sites in Escherichia coli filaments forming cells lacking DNA. J Bacteriol. 1989 Aug;171(8):4303–4314. doi: 10.1128/jb.171.8.4303-4314.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr E., Fairweather N. F., Holland I. B., Pritchard R. H. Isolation and characterisation of a strain carrying a conditional lethal mutation in the cou gene of Escherichia coli K12. Mol Gen Genet. 1979;177(1):103–112. doi: 10.1007/BF00267259. [DOI] [PubMed] [Google Scholar]
- Pruss G. J. DNA topoisomerase I mutants. Increased heterogeneity in linking number and other replicon-dependent changes in DNA supercoiling. J Mol Biol. 1985 Sep 5;185(1):51–63. doi: 10.1016/0022-2836(85)90182-2. [DOI] [PubMed] [Google Scholar]
- Ruberti I., Crescenzi F., Paolozzi L., Ghelardini P. A class of gyrB mutants, substantially unaffected in DNA topology, suppresses the Escherichia coli K12 ftsZ84 mutation. Mol Microbiol. 1991 May;5(5):1065–1072. doi: 10.1111/j.1365-2958.1991.tb01878.x. [DOI] [PubMed] [Google Scholar]
- Steck T. R., Drlica K. Bacterial chromosome segregation: evidence for DNA gyrase involvement in decatenation. Cell. 1984 Apr;36(4):1081–1088. doi: 10.1016/0092-8674(84)90058-8. [DOI] [PubMed] [Google Scholar]
- Sutcliffe J. G. Complete nucleotide sequence of the Escherichia coli plasmid pBR322. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 1):77–90. doi: 10.1101/sqb.1979.043.01.013. [DOI] [PubMed] [Google Scholar]
- Trun N. J., Gottesman S. On the bacterial cell cycle: Escherichia coli mutants with altered ploidy. Genes Dev. 1990 Dec;4(12A):2036–2047. doi: 10.1101/gad.4.12a.2036. [DOI] [PubMed] [Google Scholar]
- Tétart F., Albigot R., Conter A., Mulder E., Bouché J. P. Involvement of FtsZ in coupling of nucleoid separation with septation. Mol Microbiol. 1992 Mar;6(5):621–627. doi: 10.1111/j.1365-2958.1992.tb01509.x. [DOI] [PubMed] [Google Scholar]
- Woldringh C. L., Mulder E., Huls P. G., Vischer N. Toporegulation of bacterial division according to the nucleoid occlusion model. Res Microbiol. 1991 Feb-Apr;142(2-3):309–320. doi: 10.1016/0923-2508(91)90046-d. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- de Boer P. A., Cook W. R., Rothfield L. I. Bacterial cell division. Annu Rev Genet. 1990;24:249–274. doi: 10.1146/annurev.ge.24.120190.001341. [DOI] [PubMed] [Google Scholar]