Abstract
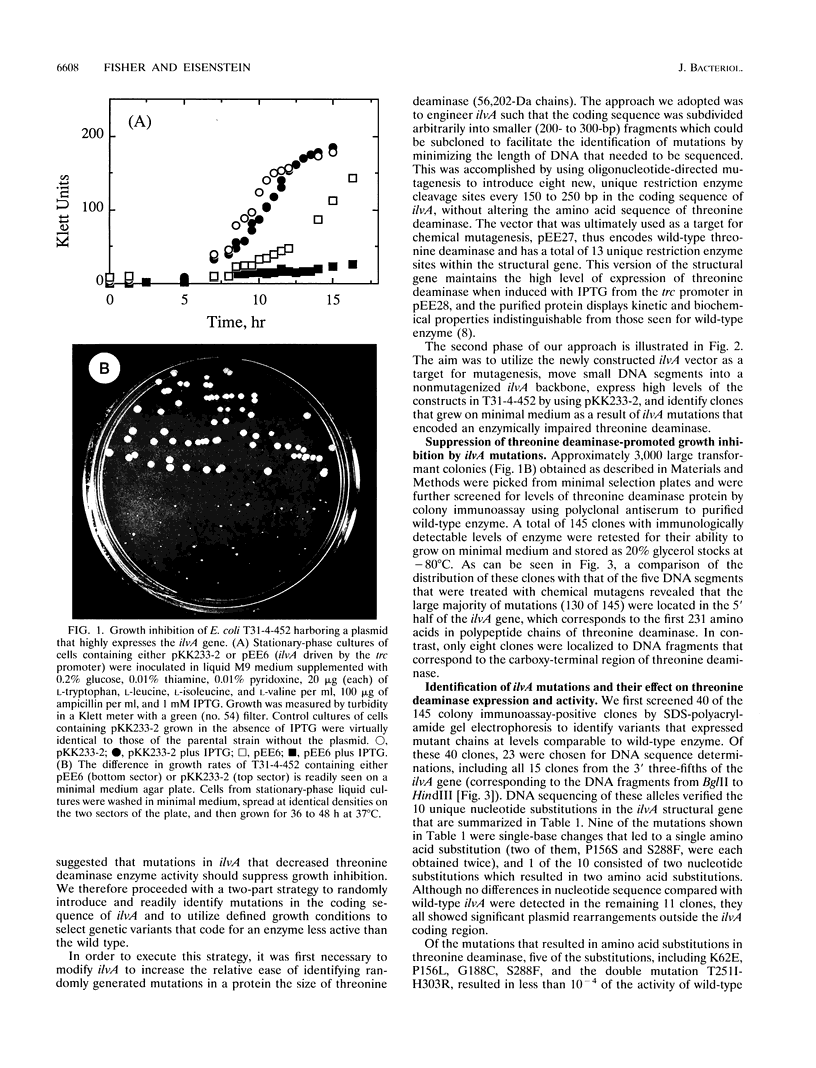
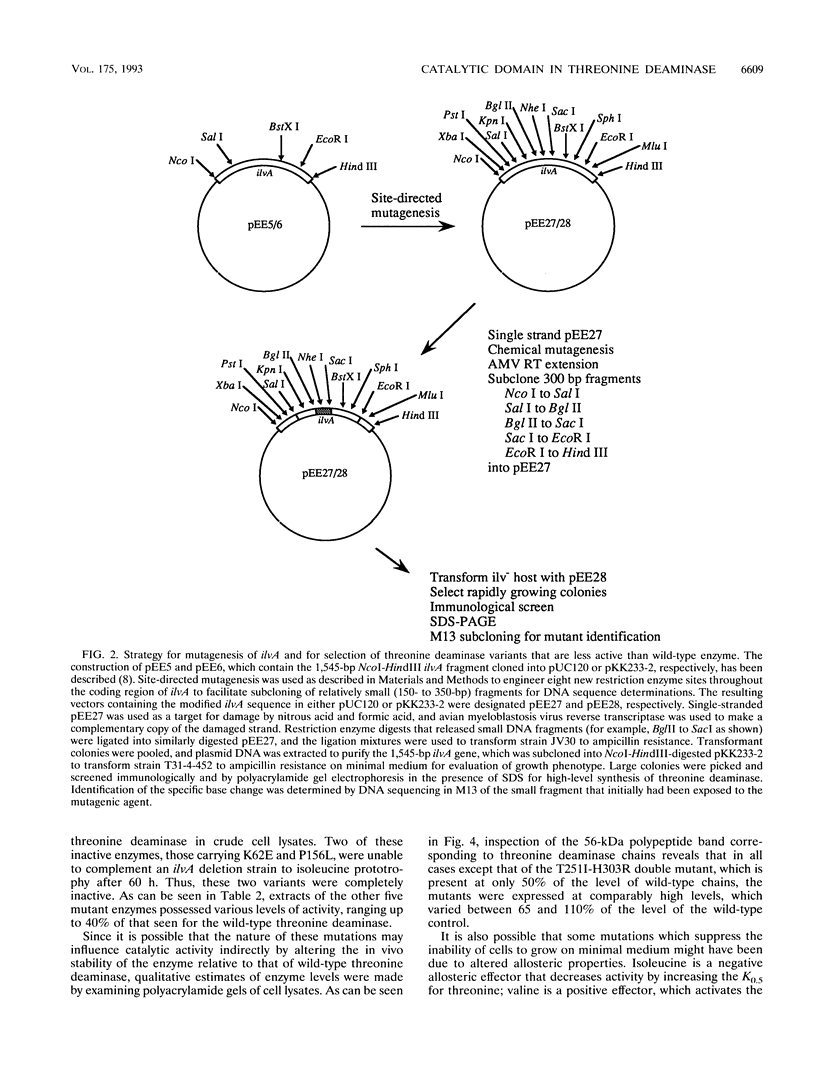
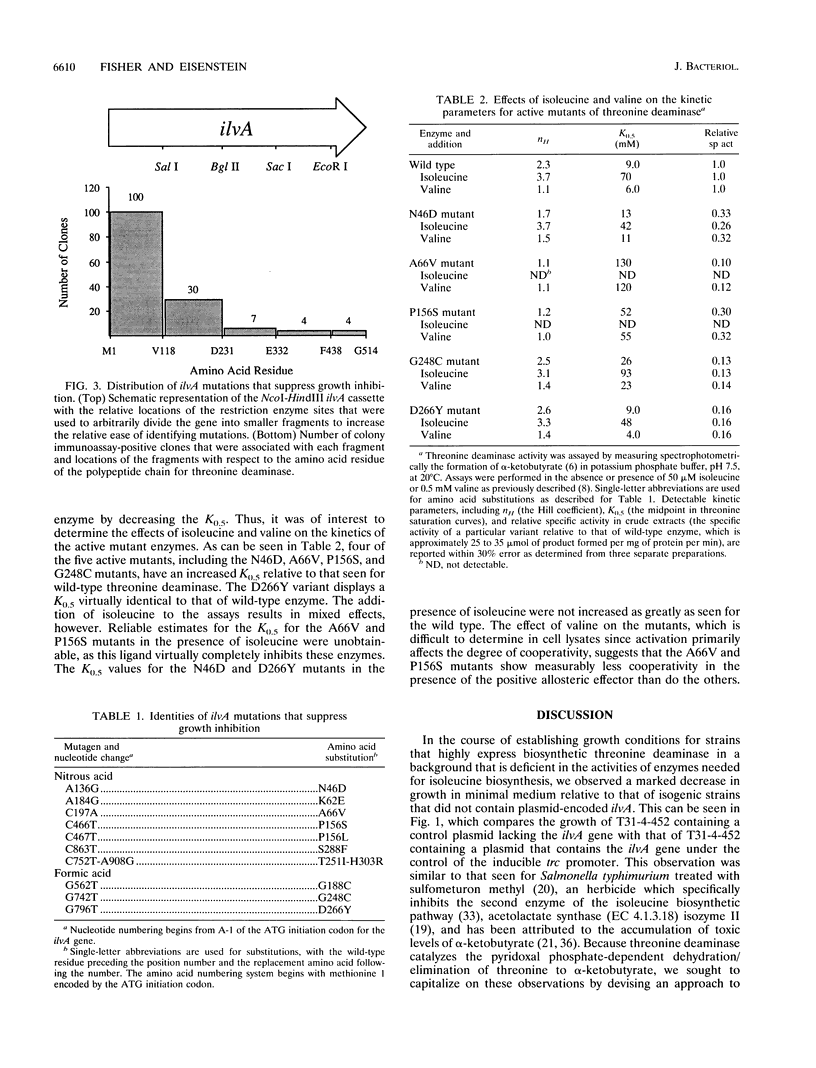
High-level expression of the regulatory enzyme threonine deaminase in Escherichia coli strains grown on minimal medium that are deficient in the activities of enzymes needed for branched-chain amino acid biosynthesis result in growth inhibition, possibly because of the accumulation of toxic levels of alpha-ketobutyrate, the product of the committed step in isoleucine biosynthesis. This condition affords a means for selecting genetic variants of threonine deaminase that are deficient in catalysis by suppression of growth inhibition. Strains harboring mutations in ilvA that decreased the catalytic activity of threonine deaminase were found to grow more rapidly than isogenic strains containing wild-type ilvA. Modification of the ilvA gene to introduce additional unique, evenly spaced restriction enzyme sites facilitated the identification of suppressor mutations by enabling small DNA fragments to be subcloned for sequencing. The 10 mutations identified in ilvA code for enzymes with significantly reduced activity relative to that of wild-type threonine deaminase. Values for their specific activities range from 40% of that displayed by wild-type enzyme to complete inactivation as evidenced by failure to complement an ilvA deletion strain to isoleucine prototrophy. Moreover, some mutant enzymes showed altered allosteric properties with respect to valine activation and isoleucine inhibition. The location of the 10 mutations in the 5' two-thirds of the ilvA gene is consistent with suggestions that threonine deaminase is organized functionally with an amino-terminal domain that is involved in catalysis and a carboxy-terminal domain that is important for regulation.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bork P., Rohde K. Sequence similarities between tryptophan synthase beta subunit and other pyridoxal-phosphate-dependent enzymes. Biochem Biophys Res Commun. 1990 Sep 28;171(3):1319–1325. doi: 10.1016/0006-291x(90)90830-g. [DOI] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Calendar R., Lindqvist B., Sironi G., Clark A. J. Characterization of REP- mutants and their interaction with P2 phage. Virology. 1970 Jan;40(1):72–83. doi: 10.1016/0042-6822(70)90380-6. [DOI] [PubMed] [Google Scholar]
- Calhoun D. H., Rimerman R. A., Hatfield G. W. Threonine deaminase from Escherichia coli. I. Purification and properties. J Biol Chem. 1973 May 25;248(10):3511–3516. [PubMed] [Google Scholar]
- Cox J. L., Cox B. J., Fidanza V., Calhoun D. H. The complete nucleotide sequence of the ilvGMEDA cluster of Escherichia coli K-12. Gene. 1987;56(2-3):185–198. doi: 10.1016/0378-1119(87)90136-3. [DOI] [PubMed] [Google Scholar]
- Davis L. A spectrophotometric method for the assay of threonine dehydratase. Anal Biochem. 1965 Jul;12(1):36–40. doi: 10.1016/0003-2697(65)90139-9. [DOI] [PubMed] [Google Scholar]
- Driver R. P., Lawther R. P. Physical analysis of deletion mutations in the ilvGEDA operon of Escherichia coli K-12. J Bacteriol. 1985 May;162(2):598–606. doi: 10.1128/jb.162.2.598-606.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenstein E. Cloning, expression, purification, and characterization of biosynthetic threonine deaminase from Escherichia coli. J Biol Chem. 1991 Mar 25;266(9):5801–5807. [PubMed] [Google Scholar]
- Fahnestock S. R., Alexander P., Nagle J., Filpula D. Gene for an immunoglobulin-binding protein from a group G streptococcus. J Bacteriol. 1986 Sep;167(3):870–880. doi: 10.1128/jb.167.3.870-880.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkes R., Niday E., Gordon J. A dot-immunobinding assay for monoclonal and other antibodies. Anal Biochem. 1982 Jan 1;119(1):142–147. doi: 10.1016/0003-2697(82)90677-7. [DOI] [PubMed] [Google Scholar]
- Holmes D. S., Quigley M. A rapid boiling method for the preparation of bacterial plasmids. Anal Biochem. 1981 Jun;114(1):193–197. doi: 10.1016/0003-2697(81)90473-5. [DOI] [PubMed] [Google Scholar]
- Hyde C. C., Ahmed S. A., Padlan E. A., Miles E. W., Davies D. R. Three-dimensional structure of the tryptophan synthase alpha 2 beta 2 multienzyme complex from Salmonella typhimurium. J Biol Chem. 1988 Nov 25;263(33):17857–17871. [PubMed] [Google Scholar]
- Kantrowitz E. R., Lipscomb W. N. Escherichia coli aspartate transcarbamoylase: the molecular basis for a concerted allosteric transition. Trends Biochem Sci. 1990 Feb;15(2):53–59. doi: 10.1016/0968-0004(90)90176-c. [DOI] [PubMed] [Google Scholar]
- Kunkel T. A., Roberts J. D., Zakour R. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- LaRossa R. A., Schloss J. V. The sulfonylurea herbicide sulfometuron methyl is an extremely potent and selective inhibitor of acetolactate synthase in Salmonella typhimurium. J Biol Chem. 1984 Jul 25;259(14):8753–8757. [PubMed] [Google Scholar]
- LaRossa R. A., Van Dyk T. K., Smulski D. R. Toxic accumulation of alpha-ketobutyrate caused by inhibition of the branched-chain amino acid biosynthetic enzyme acetolactate synthase in Salmonella typhimurium. J Bacteriol. 1987 Apr;169(4):1372–1378. doi: 10.1128/jb.169.4.1372-1378.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lawther R. P., Hatfield G. W. A site of action for tRNA mediated regulation of the ilvOEDA operon of Escherichia coli K12. Mol Gen Genet. 1978 Nov 29;167(2):227–234. doi: 10.1007/BF00266916. [DOI] [PubMed] [Google Scholar]
- Lawther R. P., Wek R. C., Lopes J. M., Pereira R., Taillon B. E., Hatfield G. W. The complete nucleotide sequence of the ilvGMEDA operon of Escherichia coli K-12. Nucleic Acids Res. 1987 Mar 11;15(5):2137–2155. doi: 10.1093/nar/15.5.2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers R. M., Lerman L. S., Maniatis T. A general method for saturation mutagenesis of cloned DNA fragments. Science. 1985 Jul 19;229(4710):242–247. doi: 10.1126/science.2990046. [DOI] [PubMed] [Google Scholar]
- Robey E. A., Wente S. R., Markby D. W., Flint A., Yang Y. R., Schachman H. K. Effect of amino acid substitutions on the catalytic and regulatory properties of aspartate transcarbamoylase. Proc Natl Acad Sci U S A. 1986 Aug;83(16):5934–5938. doi: 10.1073/pnas.83.16.5934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott J. F., Kornberg A. Purification of the rep protein of Escherichia coli. An ATPase which separates duplex DNA strands in advance of replication. J Biol Chem. 1978 May 10;253(9):3292–3297. [PubMed] [Google Scholar]
- Szentirmai A., Szentirmai M., Umbarger H. E. Isoleucine and valine metabolism of Escherichia coli. XV. Biochemical properties of mutants resistant to thiaisoleucine. J Bacteriol. 1968 May;95(5):1672–1679. doi: 10.1128/jb.95.5.1672-1679.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taillon B. E., Little R., Lawther R. P. Analysis of the functional domains of biosynthetic threonine deaminase by comparison of the amino acid sequences of three wild-type alleles to the amino acid sequence of biodegradative threonine deaminase. Gene. 1988 Mar 31;63(2):245–252. doi: 10.1016/0378-1119(88)90528-8. [DOI] [PubMed] [Google Scholar]
- Taylor J. W., Ott J., Eckstein F. The rapid generation of oligonucleotide-directed mutations at high frequency using phosphorothioate-modified DNA. Nucleic Acids Res. 1985 Dec 20;13(24):8765–8785. doi: 10.1093/nar/13.24.8765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tessman I., Fassler J. S., Bennett D. C. Relative map location of the rep and rho genes of Escherichia coli. J Bacteriol. 1982 Sep;151(3):1637–1640. doi: 10.1128/jb.151.3.1637-1640.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umbarger H. E. The origin of a useful concept--feedback inhibition. Protein Sci. 1992 Oct;1(10):1392–1395. doi: 10.1002/pro.5560011019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira J., Messing J. Production of single-stranded plasmid DNA. Methods Enzymol. 1987;153:3–11. doi: 10.1016/0076-6879(87)53044-0. [DOI] [PubMed] [Google Scholar]
- Wek R. C., Hatfield G. W. Nucleotide sequence and in vivo expression of the ilvY and ilvC genes in Escherichia coli K12. Transcription from divergent overlapping promoters. J Biol Chem. 1986 Feb 15;261(5):2441–2450. [PubMed] [Google Scholar]