Abstract
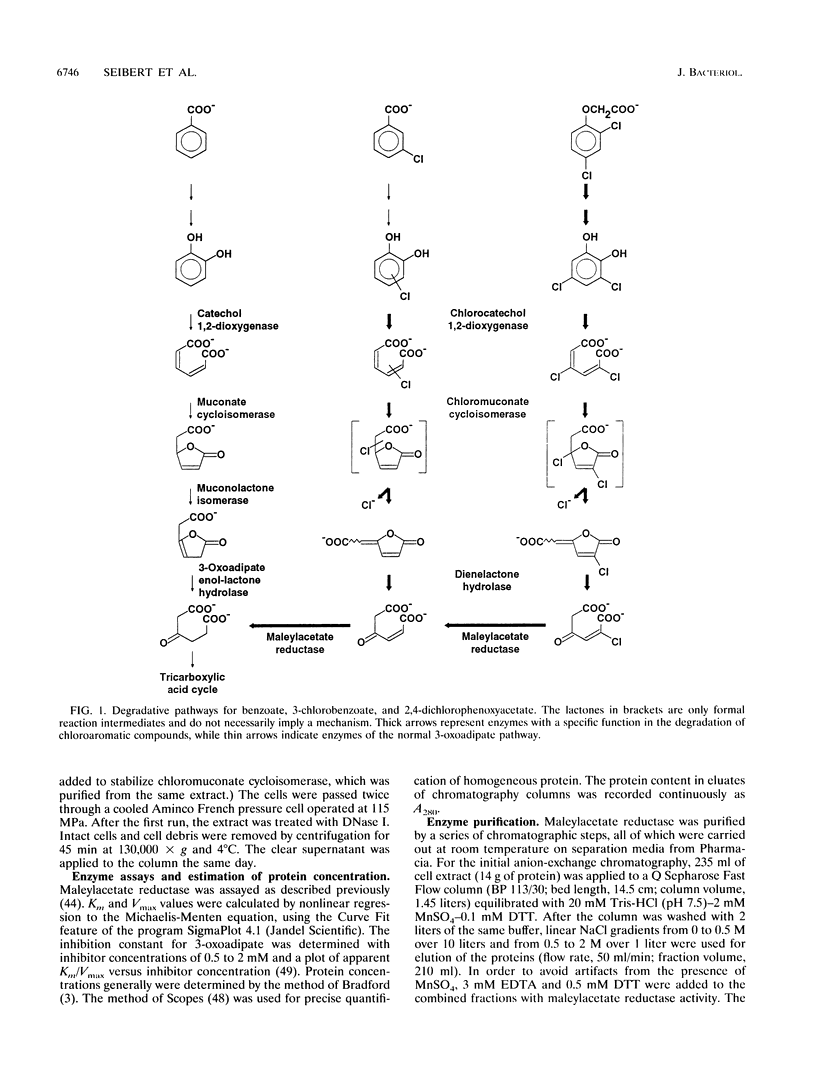
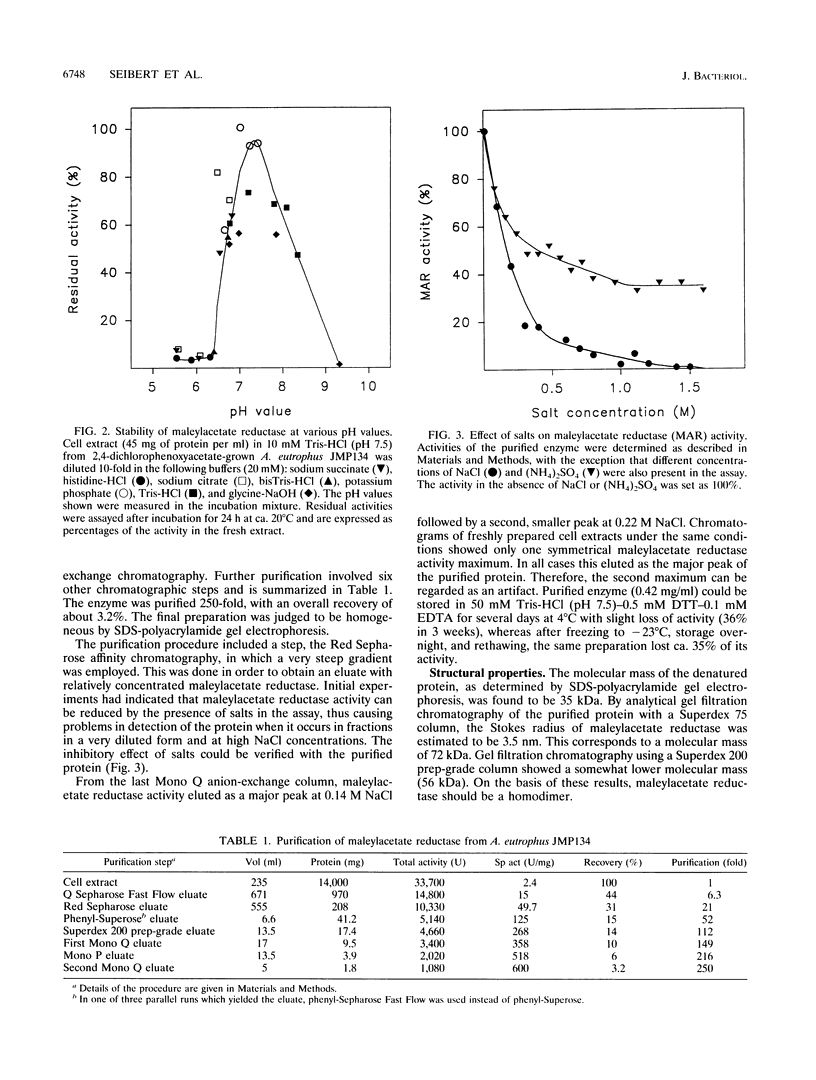
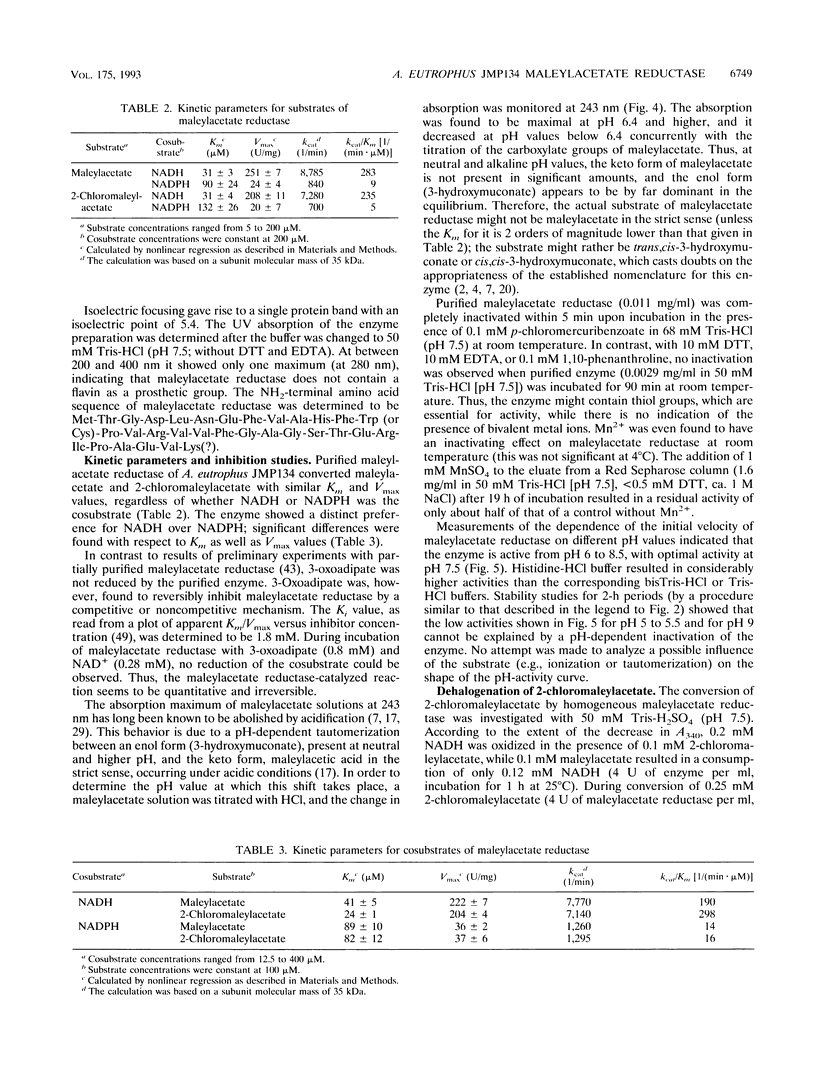
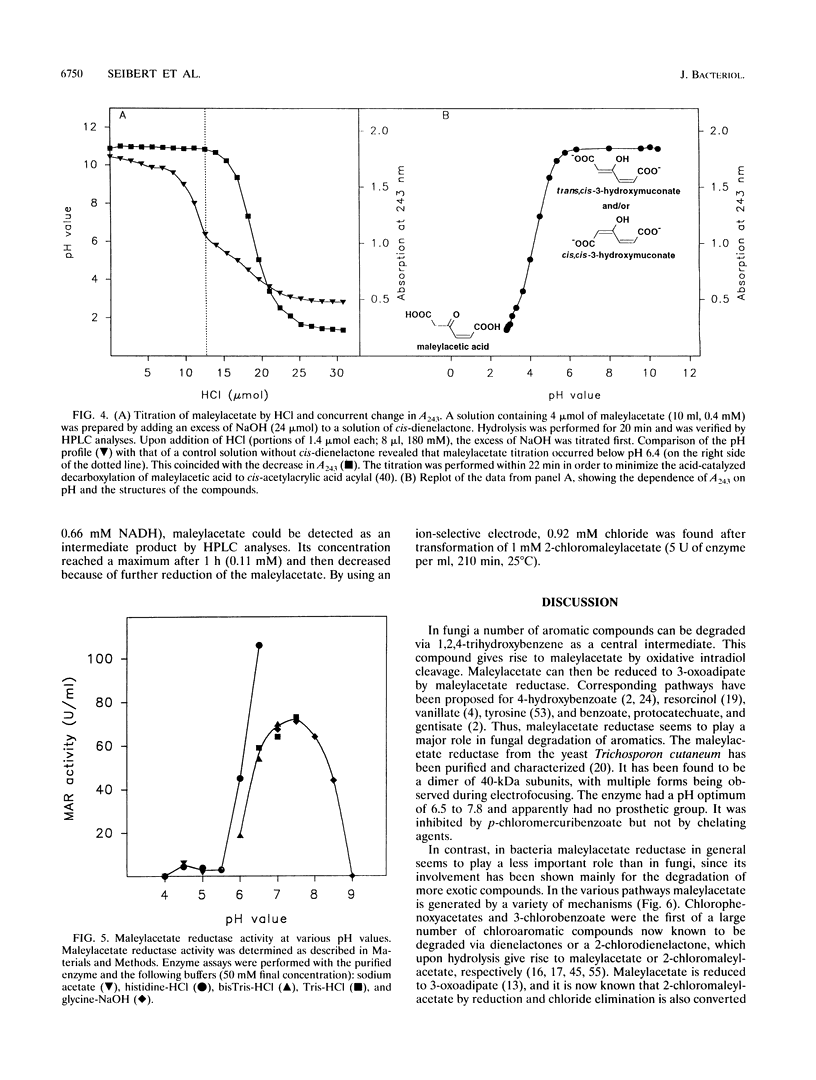
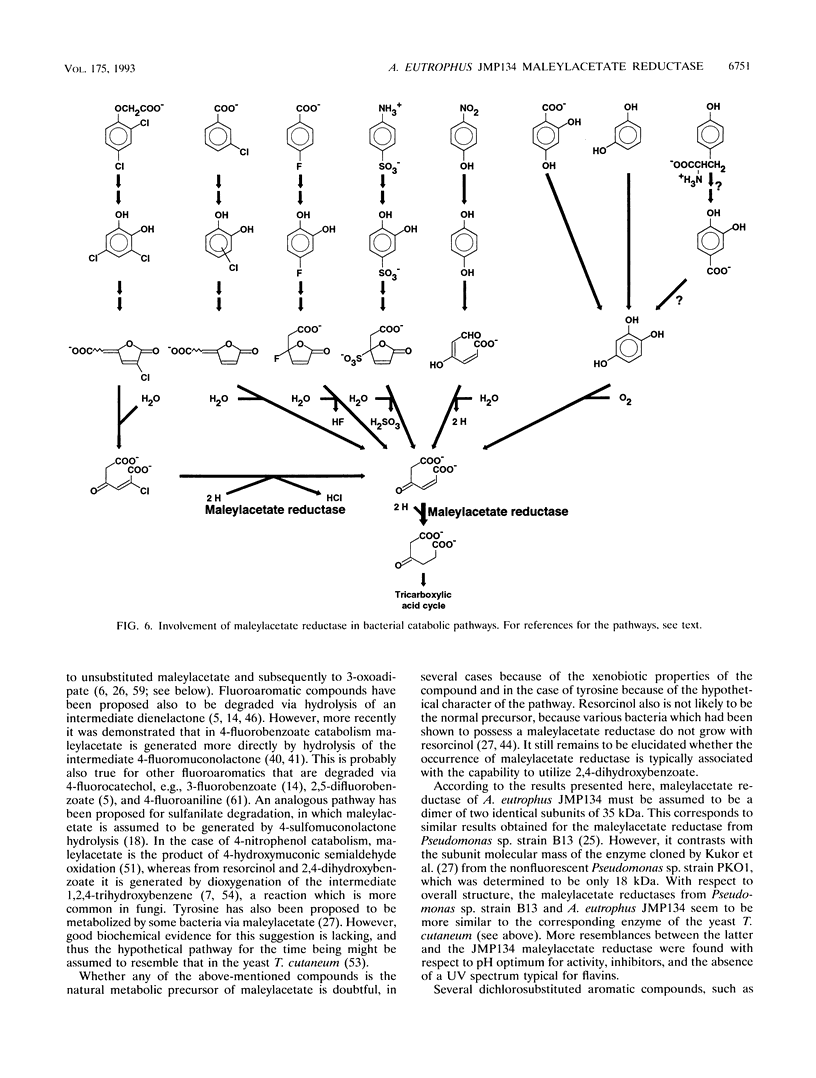
Maleylacetate reductase (EC 1.3.1.32) plays a major role in the degradation of chloroaromatic compounds by channeling maleylacetate and some of its substituted derivatives into the 3-oxoadipate pathway. The enzyme was purified to apparent homogeneity from an extract of 2,4-dichlorophenoxyacetate (2,4-D)-grown cells of Alcaligenes eutrophus JMP134. Maleylacetate reductase appears to be a dimer of two identical subunits of 35 kDa. The pI was determined to be at pH 5.4. There was no indication of a flavin prosthetic group. The enzyme was inactivated by p-chloromercuribenzoate but not by EDTA, 1,10-phenanthroline, or dithiothreitol. Maleylacetate and 2-chloromaleylacetate were converted with similar efficiencies (with NADH as cosubstrate, Km = 31 microM for each substrate and kcat = 8,785 and 7,280/min, respectively). NADH was preferred to NADPH as the cosubstrate. Upon reduction of 2-chloramaleylacetate by the purified enzyme, chloride was liberated and the resulting maleylacetate was further reduced by a second NADH. These results and the kinetic parameters suggest that the maleylacetate reductase is sufficient to channel the 2,4-D degradation intermediate 2-chloromaleylacetate into the 3-oxoadipate pathway. In a data base search the NH2-terminal sequence of maleylacetate reductase was found to be most similar to that of TfdF, a pJP4-encoded protein of as-yet-unknown function in 2,4-D degradation.
Full text
PDF




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. Basic local alignment search tool. J Mol Biol. 1990 Oct 5;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Anderson J. J., Dagley S. Catabolism of aromatic acids in Trichosporon cutaneum. J Bacteriol. 1980 Feb;141(2):534–543. doi: 10.1128/jb.141.2.534-543.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Cass A. E., Ribbons D. W., Rossiter J. T., Williams S. R. Biotransformation of aromatic compounds. Monitoring fluorinated analogues by NMR. FEBS Lett. 1987 Aug 17;220(2):353–357. doi: 10.1016/0014-5793(87)80845-1. [DOI] [PubMed] [Google Scholar]
- Chapman P. J., Ribbons D. W. Metabolism of resorcinylic compounds by bacteria: alternative pathways for resorcinol catabolism in Pseudomonas putida. J Bacteriol. 1976 Mar;125(3):985–998. doi: 10.1128/jb.125.3.985-998.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee D. K., Chakrabarty A. M. Restriction mapping of a chlorobenzoate degradative plasmid and molecular cloning of the degradative genes. Gene. 1984 Feb;27(2):173–181. doi: 10.1016/0378-1119(84)90138-0. [DOI] [PubMed] [Google Scholar]
- Chaudhry G. R., Chapalamadugu S. Biodegradation of halogenated organic compounds. Microbiol Rev. 1991 Mar;55(1):59–79. doi: 10.1128/mr.55.1.59-79.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Don R. H., Weightman A. J., Knackmuss H. J., Timmis K. N. Transposon mutagenesis and cloning analysis of the pathways for degradation of 2,4-dichlorophenoxyacetic acid and 3-chlorobenzoate in Alcaligenes eutrophus JMP134(pJP4). J Bacteriol. 1985 Jan;161(1):85–90. doi: 10.1128/jb.161.1.85-90.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorn E., Hellwig M., Reineke W., Knackmuss H. J. Isolation and characterization of a 3-chlorobenzoate degrading pseudomonad. Arch Microbiol. 1974;99(1):61–70. doi: 10.1007/BF00696222. [DOI] [PubMed] [Google Scholar]
- Dorn E., Knackmuss H. J. Chemical structure and biodegradability of halogenated aromatic compounds. Substituent effects on 1,2-dioxygenation of catechol. Biochem J. 1978 Jul 15;174(1):85–94. doi: 10.1042/bj1740085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duxbury J. M., Tiedje J. M., Alexander M., Dawson J. E. 2,4-D metabolism: enzymatic conversion of chloromaleylacetic acid to succinic acid. J Agric Food Chem. 1970 Mar-Apr;18(2):199–201. doi: 10.1021/jf60168a029. [DOI] [PubMed] [Google Scholar]
- Evans W. C., Smith B. S., Fernley H. N., Davies J. I. Bacterial metabolism of 2,4-dichlorophenoxyacetate. Biochem J. 1971 May;122(4):543–551. doi: 10.1042/bj1220543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans W. C., Smith B. S., Moss P., Fernley H. N. Bacterial metabolism of 4-chlorophenoxyacetate. Biochem J. 1971 May;122(4):509–517. doi: 10.1042/bj1220509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feigel B. J., Knackmuss H. J. Syntrophic interactions during degradation of 4-aminobenzenesulfonic acid by a two species bacterial culture. Arch Microbiol. 1993;159(2):124–130. doi: 10.1007/BF00250271. [DOI] [PubMed] [Google Scholar]
- Gaal A. B., Neujahr H. Y. Maleylacetate reductase from Trichosporon cutaneum. Biochem J. 1980 Mar 1;185(3):783–786. doi: 10.1042/bj1850783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaal A., Neujahr H. Y. Metabolism of phenol and resorcinol in Trichosporon cutaneum. J Bacteriol. 1979 Jan;137(1):13–21. doi: 10.1128/jb.137.1.13-21.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaunt J. K., Evans W. C. Metabolism of 4-chloro-2-methylphenoxyacetate by a soil pseudomonad. Ring-fission, lactonizing and delactonizing enzymes. Biochem J. 1971 May;122(4):533–542. doi: 10.1042/bj1220533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann J., Reineke W., Knackmuss H. J. Metabolism of 3-chloro-, 4-chloro-, and 3,5-dichlorobenzoate by a pseudomonad. Appl Environ Microbiol. 1979 Mar;37(3):421–428. doi: 10.1128/aem.37.3.421-428.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Häggblom M. M. Microbial breakdown of halogenated aromatic pesticides and related compounds. FEMS Microbiol Rev. 1992 Sep;9(1):29–71. doi: 10.1111/j.1574-6968.1992.tb05823.x. [DOI] [PubMed] [Google Scholar]
- Kaschabek S. R., Reineke W. Maleylacetate reductase of Pseudomonas sp. strain B13: dechlorination of chloromaleylacetates, metabolites in the degradation of chloroaromatic compounds. Arch Microbiol. 1992;158(6):412–417. doi: 10.1007/BF00276301. [DOI] [PubMed] [Google Scholar]
- Kukor J. J., Olsen R. H., Siak J. S. Recruitment of a chromosomally encoded maleylacetate reductase for degradation of 2,4-dichlorophenoxyacetic acid by plasmid pJP4. J Bacteriol. 1989 Jun;171(6):3385–3390. doi: 10.1128/jb.171.6.3385-3390.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Merril C. R., Goldman D., Sedman S. A., Ebert M. H. Ultrasensitive stain for proteins in polyacrylamide gels shows regional variation in cerebrospinal fluid proteins. Science. 1981 Mar 27;211(4489):1437–1438. doi: 10.1126/science.6162199. [DOI] [PubMed] [Google Scholar]
- Perkins E. J., Gordon M. P., Caceres O., Lurquin P. F. Organization and sequence analysis of the 2,4-dichlorophenol hydroxylase and dichlorocatechol oxidative operons of plasmid pJP4. J Bacteriol. 1990 May;172(5):2351–2359. doi: 10.1128/jb.172.5.2351-2359.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reineke W., Knackmuss H. J. Microbial degradation of haloaromatics. Annu Rev Microbiol. 1988;42:263–287. doi: 10.1146/annurev.mi.42.100188.001403. [DOI] [PubMed] [Google Scholar]
- Sander P., Wittich R. M., Fortnagel P., Wilkes H., Francke W. Degradation of 1,2,4-trichloro- and 1,2,4,5-tetrachlorobenzene by pseudomonas strains. Appl Environ Microbiol. 1991 May;57(5):1430–1440. doi: 10.1128/aem.57.5.1430-1440.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlömann M., Fischer P., Schmidt E., Knackmuss H. J. Enzymatic formation, stability, and spontaneous reactions of 4-fluoromuconolactone, a metabolite of the bacterial degradation of 4-fluorobenzoate. J Bacteriol. 1990 Sep;172(9):5119–5129. doi: 10.1128/jb.172.9.5119-5129.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlömann M., Ngai K. L., Ornston L. N., Knackmuss H. J. Dienelactone hydrolase from Pseudomonas cepacia. J Bacteriol. 1993 May;175(10):2994–3001. doi: 10.1128/jb.175.10.2994-3001.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlömann M., Schmidt E., Knackmuss H. J. Different types of dienelactone hydrolase in 4-fluorobenzoate-utilizing bacteria. J Bacteriol. 1990 Sep;172(9):5112–5118. doi: 10.1128/jb.172.9.5112-5118.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt E., Knackmuss H. J. Chemical structure and biodegradability of halogenated aromatic compounds. Conversion of chlorinated muconic acids into maleoylacetic acid. Biochem J. 1980 Oct 15;192(1):339–347. doi: 10.1042/bj1920339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber A., Hellwig M., Dorn E., Reineke W., Knackmuss H. J. Critical Reactions in Fluorobenzoic Acid Degradation by Pseudomonas sp. B13. Appl Environ Microbiol. 1980 Jan;39(1):58–67. doi: 10.1128/aem.39.1.58-67.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scopes R. K. Measurement of protein by spectrophotometry at 205 nm. Anal Biochem. 1974 May;59(1):277–282. doi: 10.1016/0003-2697(74)90034-7. [DOI] [PubMed] [Google Scholar]
- Spain J. C., Gibson D. T. Pathway for Biodegradation of p-Nitrophenol in a Moraxella sp. Appl Environ Microbiol. 1991 Mar;57(3):812–819. doi: 10.1128/aem.57.3.812-819.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spain J. C., Nishino S. F. Degradation of 1,4-dichlorobenzene by a Pseudomonas sp. Appl Environ Microbiol. 1987 May;53(5):1010–1019. doi: 10.1128/aem.53.5.1010-1019.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparnins V. L., Burbee D. G., Dagley S. Catabolism of L-tyrosine in Trichosporon cutaneum. J Bacteriol. 1979 May;138(2):425–430. doi: 10.1128/jb.138.2.425-430.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolz A., Knackmuss H. J. Degradation of 2,4-dihydroxybenzoate by Pseudomonas sp. BN9. FEMS Microbiol Lett. 1993 Apr 1;108(2):219–224. doi: 10.1111/j.1574-6968.1993.tb06102.x. [DOI] [PubMed] [Google Scholar]
- Vollmer M. D., Stadler-Fritzsche K., Schlömann M. Conversion of 2-chloromaleylacetate in Alcaligenes eutrophus JMP134. Arch Microbiol. 1993;159(2):182–188. doi: 10.1007/BF00250280. [DOI] [PubMed] [Google Scholar]
- Zeyer J., Wasserfallen A., Timmis K. N. Microbial mineralization of ring-substituted anilines through an ortho-cleavage pathway. Appl Environ Microbiol. 1985 Aug;50(2):447–453. doi: 10.1128/aem.50.2.447-453.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Meer J. R., Eggen R. I., Zehnder A. J., de Vos W. M. Sequence analysis of the Pseudomonas sp. strain P51 tcb gene cluster, which encodes metabolism of chlorinated catechols: evidence for specialization of catechol 1,2-dioxygenases for chlorinated substrates. J Bacteriol. 1991 Apr;173(8):2425–2434. doi: 10.1128/jb.173.8.2425-2434.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Meer J. R., de Vos W. M., Harayama S., Zehnder A. J. Molecular mechanisms of genetic adaptation to xenobiotic compounds. Microbiol Rev. 1992 Dec;56(4):677–694. doi: 10.1128/mr.56.4.677-694.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Meer J. R., van Neerven A. R., de Vries E. J., de Vos W. M., Zehnder A. J. Cloning and characterization of plasmid-encoded genes for the degradation of 1,2-dichloro-, 1,4-dichloro-, and 1,2,4-trichlorobenzene of Pseudomonas sp. strain P51. J Bacteriol. 1991 Jan;173(1):6–15. doi: 10.1128/jb.173.1.6-15.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]



