Abstract
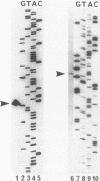
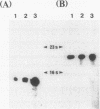
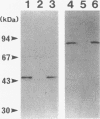
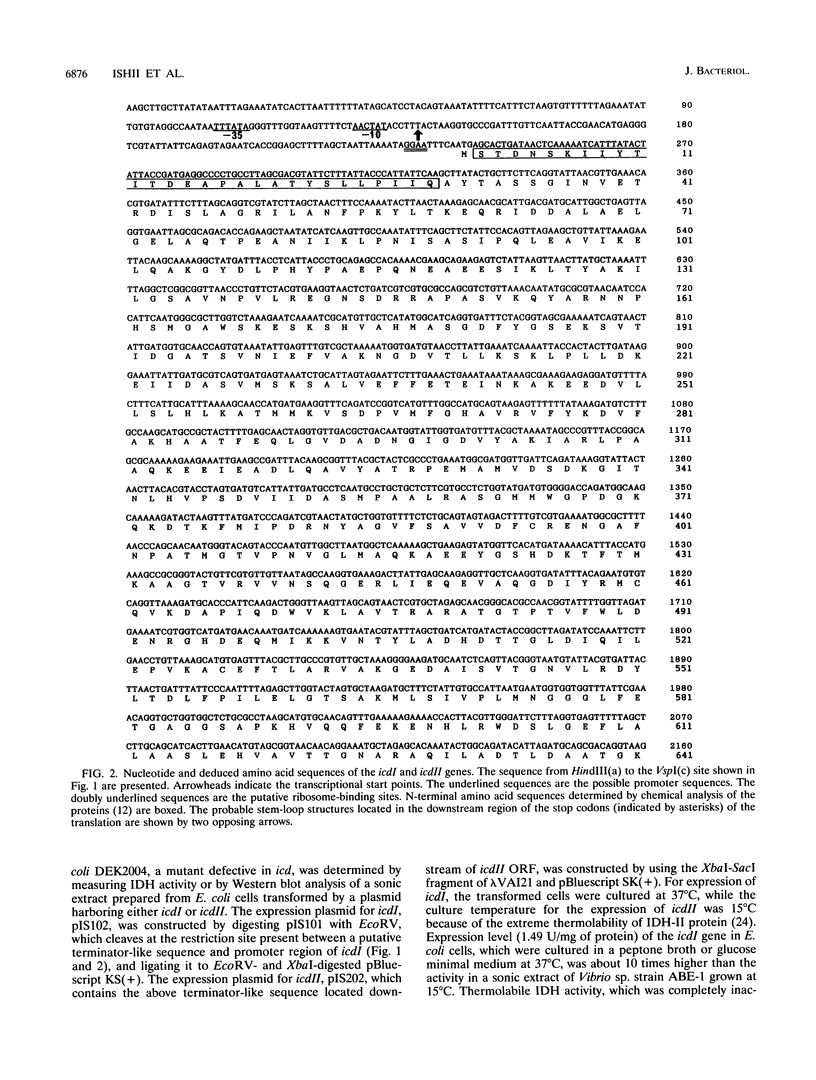
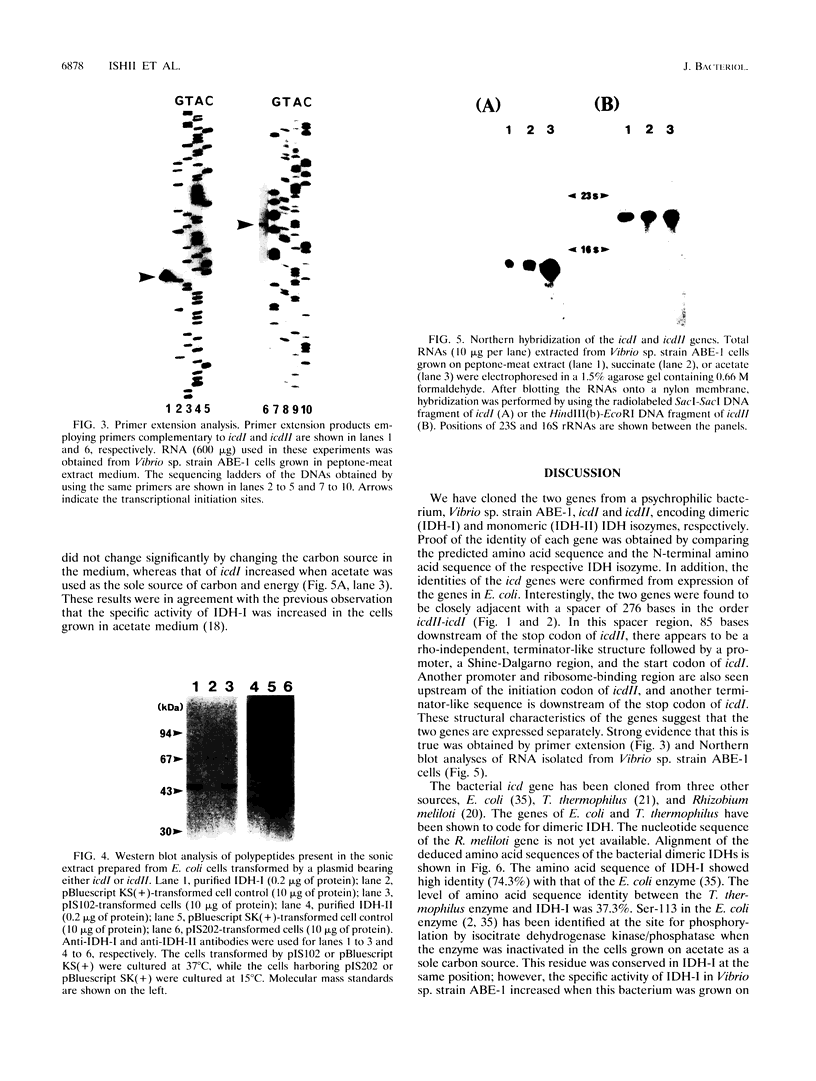
The genes coding for two structurally different isocitrate dehydrogenase isozymes (IDH-I and IDH-II) of a psychrophilic bacterium, Vibrio sp. strain ABE-1, were cloned and sequenced. Open reading frames of the genes (icdI and icdII) are 1,248 and 2,229 bp in length, respectively. The amino acid sequences predicted from the open reading frames of icdI and icdII corresponded to the N-terminal amino acid sequences of the purified IDH-I and IDH-II, respectively. No homology was found between the deduced amino acid sequences of the isozymes; however, the IDH-I, a dimeric enzyme, had a high amino acid sequence identity (74.3%) to the Escherichia coli IDH. The deduced amino acid sequence of the IDH-II, a monomeric enzyme, was not related to any known sequence. However, the IDH-II had an amino acid sequence homologous to that of a cyanogen bromide-cleaved peptide containing a putative active-site methionyl residue of the monomeric IDH of Azotobacter vinelandii. The two genes (icdlI and icdII) were found to be tandemly located in the same orientation. Northern (RNA) blot analyses showed that the two genes are transcribed independently. Primer extension experiments located single transcriptional start sites 39 and 96 bp upstream of the start codons of icdI and icdII, respectively. The amount of icdI transcript but not icdII increased when Vibrio sp. strain ABE-1 cells were cultured in acetate minimal medium.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bedbrook J. R., Jones J., O'Dell M., Thompson R. D., Flavell R. B. A molecular description of telometic heterochromatin in secale species. Cell. 1980 Feb;19(2):545–560. doi: 10.1016/0092-8674(80)90529-2. [DOI] [PubMed] [Google Scholar]
- Borthwick A. C., Holms W. H., Nimmo H. G. Amino acid sequence round the site of phosphorylation in isocitrate dehydrogenase from Escherichia coli ML308. FEBS Lett. 1984 Aug 20;174(1):112–115. doi: 10.1016/0014-5793(84)81087-x. [DOI] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Burke W. F., Johanson R. A., Reeves H. C. NADP+-specific isocitrate dehydrogenase of Escherichia coli. II. Subunit structure. Biochim Biophys Acta. 1974 Jun 7;351(2):333–340. doi: 10.1016/0005-2795(74)90196-2. [DOI] [PubMed] [Google Scholar]
- Case C. C., Roels S. M., González J. E., Simons E. L., Simons R. W. Analysis of the promoters and transcripts involved in IS10 anti-sense RNA control. Gene. 1988 Dec 10;72(1-2):219–236. doi: 10.1016/0378-1119(88)90147-3. [DOI] [PubMed] [Google Scholar]
- Chung A. E., Braginski J. E. Isocitrate dehydrogenase from Rhodopseudomonas spheroides: purification and characterization. Arch Biochem Biophys. 1972 Nov;153(1):357–367. doi: 10.1016/0003-9861(72)90456-0. [DOI] [PubMed] [Google Scholar]
- Chung A. E., Franzen J. S. Oxidized triphosphopyridine nucleotide specific isocitrate dehydrogenase from Azotobacter vinelandii. Isolation and characterization. Biochemistry. 1969 Aug;8(8):3175–3184. doi: 10.1021/bi00836a007. [DOI] [PubMed] [Google Scholar]
- Cupp J. R., McAlister-Henn L. Cloning and characterization of the gene encoding the IDH1 subunit of NAD(+)-dependent isocitrate dehydrogenase from Saccharomyces cerevisiae. J Biol Chem. 1992 Aug 15;267(23):16417–16423. [PubMed] [Google Scholar]
- Edwards D. J., Heinrikson R. L., Chung A. E. Triphosphopyridine nucleotide specific isocitrate dehydrogenase from Azotobacter vinelandii. Alkylation of a specific methionine residue and amino acid sequence of the peptide containing this residue. Biochemistry. 1974 Feb 12;13(4):677–683. doi: 10.1021/bi00701a007. [DOI] [PubMed] [Google Scholar]
- Fukunaga N., Imagawa S., Sahara T., Ishii A., Suzuki M. Purification and characterization of monomeric isocitrate dehydrogenase with NADP(+)-specificity from Vibrio parahaemolyticus Y-4. J Biochem. 1992 Dec;112(6):849–855. doi: 10.1093/oxfordjournals.jbchem.a123988. [DOI] [PubMed] [Google Scholar]
- Hattori M., Sakaki Y. Dideoxy sequencing method using denatured plasmid templates. Anal Biochem. 1986 Feb 1;152(2):232–238. doi: 10.1016/0003-2697(86)90403-3. [DOI] [PubMed] [Google Scholar]
- Henikoff S. Unidirectional digestion with exonuclease III in DNA sequence analysis. Methods Enzymol. 1987;155:156–165. doi: 10.1016/0076-6879(87)55014-5. [DOI] [PubMed] [Google Scholar]
- Howard R. L., Becker R. R. Isolation and some properties of the triphosphopyridine nucleotide isocitrate dehydrogenase from Bacillus stearothermophilus. J Biol Chem. 1970 Jun;245(12):3186–3194. [PubMed] [Google Scholar]
- Hurley J. H., Thorsness P. E., Ramalingam V., Helmers N. H., Koshland D. E., Jr, Stroud R. M. Structure of a bacterial enzyme regulated by phosphorylation, isocitrate dehydrogenase. Proc Natl Acad Sci U S A. 1989 Nov;86(22):8635–8639. doi: 10.1073/pnas.86.22.8635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii A., Ochiai T., Imagawa S., Fukunaga N., Sasaki S., Minowa O., Mizuno Y., Shiokawa H. Isozymes of isocitrate dehydrogenase from an obligately psychrophilic bacterium, Vibrio sp. strain ABE-1: purification, and modulation of activities by growth conditions. J Biochem. 1987 Dec;102(6):1489–1498. doi: 10.1093/oxfordjournals.jbchem.a122196. [DOI] [PubMed] [Google Scholar]
- Leyland M. L., Kelly D. J. Purification and characterization of a monomeric isocitrate dehydrogenase with dual coenzyme specificity from the photosynthetic bacterium Rhodomicrobium vannielii. Eur J Biochem. 1991 Nov 15;202(1):85–93. doi: 10.1111/j.1432-1033.1991.tb16347.x. [DOI] [PubMed] [Google Scholar]
- McDermott T. R., Kahn M. L. Cloning and mutagenesis of the Rhizobium meliloti isocitrate dehydrogenase gene. J Bacteriol. 1992 Jul;174(14):4790–4797. doi: 10.1128/jb.174.14.4790-4797.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazaki K., Eguchi H., Yamagishi A., Wakagi T., Oshima T. Molecular cloning of the isocitrate dehydrogenase gene of an extreme thermophile, Thermus thermophilus HB8. Appl Environ Microbiol. 1992 Jan;58(1):93–98. doi: 10.1128/aem.58.1.93-98.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neidhardt F. C., Bloch P. L., Smith D. F. Culture medium for enterobacteria. J Bacteriol. 1974 Sep;119(3):736–747. doi: 10.1128/jb.119.3.736-747.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nègre D., Cortay J. C., Galinier A., Sauve P., Cozzone A. J. Specific interactions between the IclR repressor of the acetate operon of Escherichia coli and its operator. J Mol Biol. 1992 Nov 5;228(1):23–29. doi: 10.1016/0022-2836(92)90488-6. [DOI] [PubMed] [Google Scholar]
- Ochiai T., Fukunaga N., Sasaki S. Purification and some properties of two NADP+-specific isocitrate dehydrogenases from an obligately psychrophilic marine bacterium, Vibrio sp., strain ABE-1. J Biochem. 1979 Aug;86(2):377–384. doi: 10.1093/oxfordjournals.jbchem.a132536. [DOI] [PubMed] [Google Scholar]
- Qoronfleh M. W., Debouck C., Keller J. Identification and characterization of novel low-temperature-inducible promoters of Escherichia coli. J Bacteriol. 1992 Dec;174(24):7902–7909. doi: 10.1128/jb.174.24.7902-7909.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg M., Court D. Regulatory sequences involved in the promotion and termination of RNA transcription. Annu Rev Genet. 1979;13:319–353. doi: 10.1146/annurev.ge.13.120179.001535. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shine J., Dalgarno L. The 3'-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc Natl Acad Sci U S A. 1974 Apr;71(4):1342–1346. doi: 10.1073/pnas.71.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Short J. M., Fernandez J. M., Sorge J. A., Huse W. D. Lambda ZAP: a bacteriophage lambda expression vector with in vivo excision properties. Nucleic Acids Res. 1988 Aug 11;16(15):7583–7600. doi: 10.1093/nar/16.15.7583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soundar S., Colman R. F. Identification of metal-isocitrate binding site of pig heart NADP-specific isocitrate dehydrogenase by affinity cleavage of the enzyme by Fe(2+)-isocitrate. J Biol Chem. 1993 Mar 5;268(7):5264–5271. [PubMed] [Google Scholar]
- Staden R. An interactive graphics program for comparing and aligning nucleic acid and amino acid sequences. Nucleic Acids Res. 1982 May 11;10(9):2951–2961. doi: 10.1093/nar/10.9.2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorsness P. E., Koshland D. E., Jr Inactivation of isocitrate dehydrogenase by phosphorylation is mediated by the negative charge of the phosphate. J Biol Chem. 1987 Aug 5;262(22):10422–10425. [PubMed] [Google Scholar]
- Wilbur W. J., Lipman D. J. Rapid similarity searches of nucleic acid and protein data banks. Proc Natl Acad Sci U S A. 1983 Feb;80(3):726–730. doi: 10.1073/pnas.80.3.726. [DOI] [PMC free article] [PubMed] [Google Scholar]