Abstract
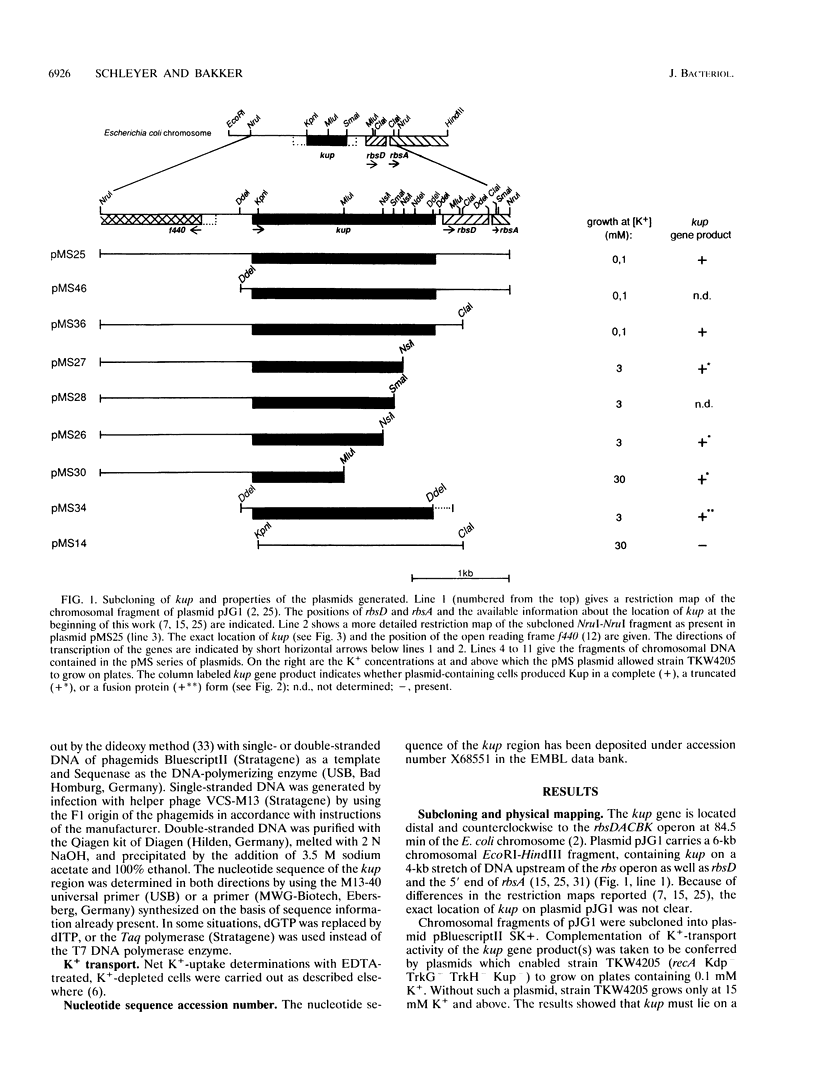
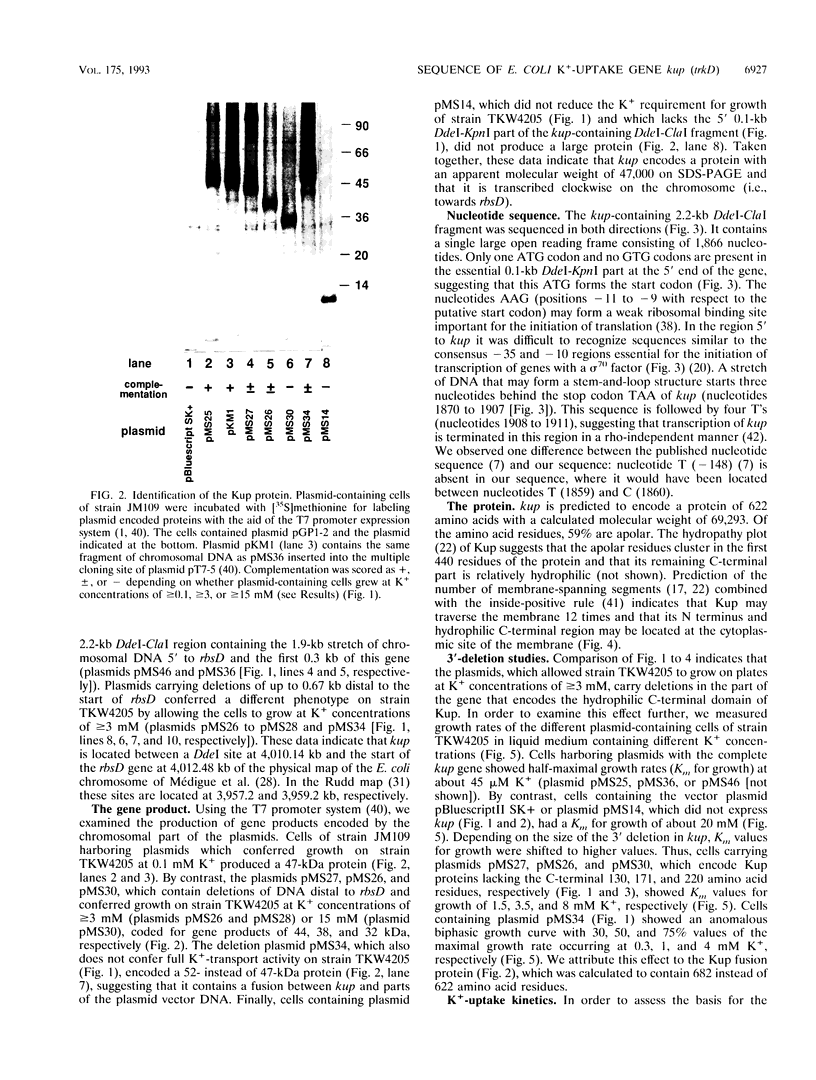
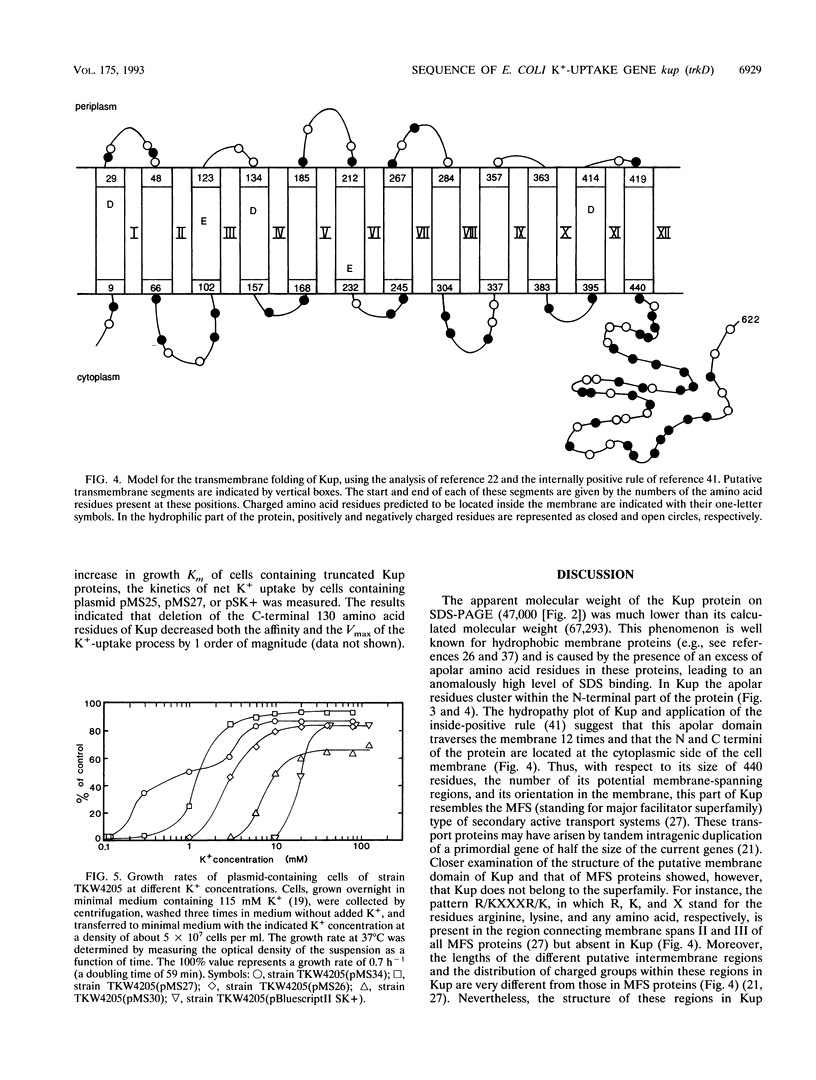
The kup (formerly trkD) gene from Escherichia coli encodes a minor K(+)-uptake system. The gene is located just upstream of the rbsDACBK operon at 84.5 min on the chromosome and is transcribed clockwise. kup codes for a 69-kDa protein, which may be composed of two domains. The first 440 amino acid residues appear to form an integral membrane protein that might traverse the cell membrane 12 times. The C-terminal 182 amino acid residues are predicted to form a hydrophilic domain located at the cytoplasmic side of the membrane. Deletion studies from the 3' end of kup showed that removal of almost the complete hydrophilic domain of the protein reduced, but did not abolish, K(+)-uptake activity.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bachmann B. J. Linkage map of Escherichia coli K-12, edition 8. Microbiol Rev. 1990 Jun;54(2):130–197. doi: 10.1128/mr.54.2.130-197.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakker E. P., Mangerich W. E. Interconversion of components of the bacterial proton motive force by electrogenic potassium transport. J Bacteriol. 1981 Sep;147(3):820–826. doi: 10.1128/jb.147.3.820-826.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell A. W., Buckel S. D., Groarke J. M., Hope J. N., Kingsley D. H., Hermodson M. A. The nucleotide sequences of the rbsD, rbsA, and rbsC genes of Escherichia coli K12. J Biol Chem. 1986 Jun 15;261(17):7652–7658. [PubMed] [Google Scholar]
- Booth I. R. Regulation of cytoplasmic pH in bacteria. Microbiol Rev. 1985 Dec;49(4):359–378. doi: 10.1128/mr.49.4.359-378.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossemeyer D., Schlösser A., Bakker E. P. Specific cesium transport via the Escherichia coli Kup (TrkD) K+ uptake system. J Bacteriol. 1989 Apr;171(4):2219–2221. doi: 10.1128/jb.171.4.2219-2221.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burland V., Plunkett G., 3rd, Daniels D. L., Blattner F. R. DNA sequence and analysis of 136 kilobases of the Escherichia coli genome: organizational symmetry around the origin of replication. Genomics. 1993 Jun;16(3):551–561. doi: 10.1006/geno.1993.1230. [DOI] [PubMed] [Google Scholar]
- Culham D. E., Lasby B., Marangoni A. G., Milner J. L., Steer B. A., van Nues R. W., Wood J. M. Isolation and sequencing of Escherichia coli gene proP reveals unusual structural features of the osmoregulatory proline/betaine transporter, ProP. J Mol Biol. 1993 Jan 5;229(1):268–276. doi: 10.1006/jmbi.1993.1030. [DOI] [PubMed] [Google Scholar]
- Dinnbier U., Limpinsel E., Schmid R., Bakker E. P. Transient accumulation of potassium glutamate and its replacement by trehalose during adaptation of growing cells of Escherichia coli K-12 to elevated sodium chloride concentrations. Arch Microbiol. 1988;150(4):348–357. doi: 10.1007/BF00408306. [DOI] [PubMed] [Google Scholar]
- Dosch D. C., Helmer G. L., Sutton S. H., Salvacion F. F., Epstein W. Genetic analysis of potassium transport loci in Escherichia coli: evidence for three constitutive systems mediating uptake potassium. J Bacteriol. 1991 Jan;173(2):687–696. doi: 10.1128/jb.173.2.687-696.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg D., Schwarz E., Komaromy M., Wall R. Analysis of membrane and surface protein sequences with the hydrophobic moment plot. J Mol Biol. 1984 Oct 15;179(1):125–142. doi: 10.1016/0022-2836(84)90309-7. [DOI] [PubMed] [Google Scholar]
- Epstein W., Kim B. S. Potassium transport loci in Escherichia coli K-12. J Bacteriol. 1971 Nov;108(2):639–644. doi: 10.1128/jb.108.2.639-644.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harley C. B., Reynolds R. P. Analysis of E. coli promoter sequences. Nucleic Acids Res. 1987 Mar 11;15(5):2343–2361. doi: 10.1093/nar/15.5.2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson P. J., Maiden M. C. Homologous sugar transport proteins in Escherichia coli and their relatives in both prokaryotes and eukaryotes. Philos Trans R Soc Lond B Biol Sci. 1990 Jan 30;326(1236):391–410. doi: 10.1098/rstb.1990.0020. [DOI] [PubMed] [Google Scholar]
- Kyte J., Doolittle R. F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982 May 5;157(1):105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Lamark T., Kaasen I., Eshoo M. W., Falkenberg P., McDougall J., Strøm A. R. DNA sequence and analysis of the bet genes encoding the osmoregulatory choline-glycine betaine pathway of Escherichia coli. Mol Microbiol. 1991 May;5(5):1049–1064. doi: 10.1111/j.1365-2958.1991.tb01877.x. [DOI] [PubMed] [Google Scholar]
- Lengeler J. W., Titgemeyer F., Vogler A. P., Wöhrl B. M. Structures and homologies of carbohydrate: phosphotransferase system (PTS) proteins. Philos Trans R Soc Lond B Biol Sci. 1990 Jan 30;326(1236):489–504. doi: 10.1098/rstb.1990.0027. [DOI] [PubMed] [Google Scholar]
- Lopilato J. E., Garwin J. L., Emr S. D., Silhavy T. J., Beckwith J. R. D-ribose metabolism in Escherichia coli K-12: genetics, regulation, and transport. J Bacteriol. 1984 May;158(2):665–673. doi: 10.1128/jb.158.2.665-673.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marger M. D., Saier M. H., Jr A major superfamily of transmembrane facilitators that catalyse uniport, symport and antiport. Trends Biochem Sci. 1993 Jan;18(1):13–20. doi: 10.1016/0968-0004(93)90081-w. [DOI] [PubMed] [Google Scholar]
- Munro A. W., Ritchie G. Y., Lamb A. J., Douglas R. M., Booth I. R. The cloning and DNA sequence of the gene for the glutathione-regulated potassium-efflux system KefC of Escherichia coli. Mol Microbiol. 1991 Mar;5(3):607–616. doi: 10.1111/j.1365-2958.1991.tb00731.x. [DOI] [PubMed] [Google Scholar]
- Médigue C., Bouché J. P., Hénaut A., Danchin A. Mapping of sequenced genes (700 kbp) in the restriction map of the Escherichia coli chromosome. Mol Microbiol. 1990 Feb;4(2):169–187. doi: 10.1111/j.1365-2958.1990.tb00585.x. [DOI] [PubMed] [Google Scholar]
- Rhoads D. B., Waters F. B., Epstein W. Cation transport in Escherichia coli. VIII. Potassium transport mutants. J Gen Physiol. 1976 Mar;67(3):325–341. doi: 10.1085/jgp.67.3.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlösser A., Kluttig S., Hamann A., Bakker E. P. Subcloning, nucleotide sequence, and expression of trkG, a gene that encodes an integral membrane protein involved in potassium uptake via the Trk system of Escherichia coli. J Bacteriol. 1991 May;173(10):3170–3176. doi: 10.1128/jb.173.10.3170-3176.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schägger H., von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem. 1987 Nov 1;166(2):368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- Shine J., Dalgarno L. Determinant of cistron specificity in bacterial ribosomes. Nature. 1975 Mar 6;254(5495):34–38. doi: 10.1038/254034a0. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- von Hippel P. H., Bear D. G., Morgan W. D., McSwiggen J. A. Protein-nucleic acid interactions in transcription: a molecular analysis. Annu Rev Biochem. 1984;53:389–446. doi: 10.1146/annurev.bi.53.070184.002133. [DOI] [PubMed] [Google Scholar]



