Abstract
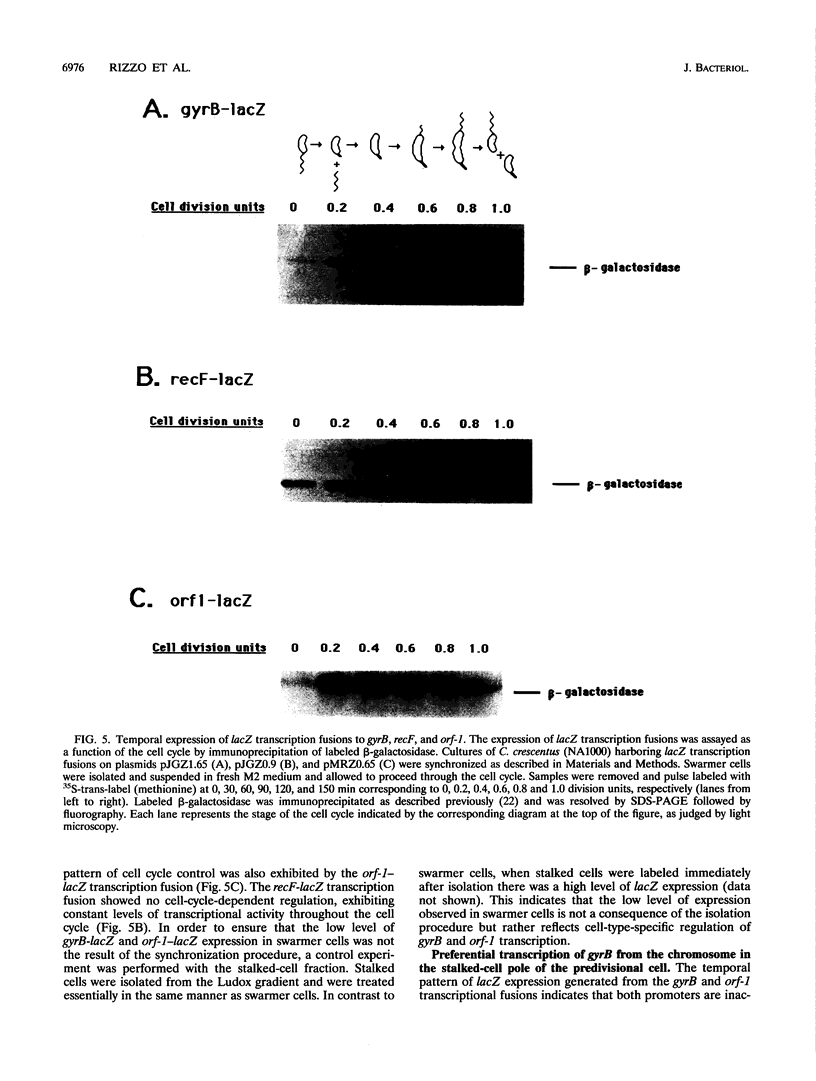
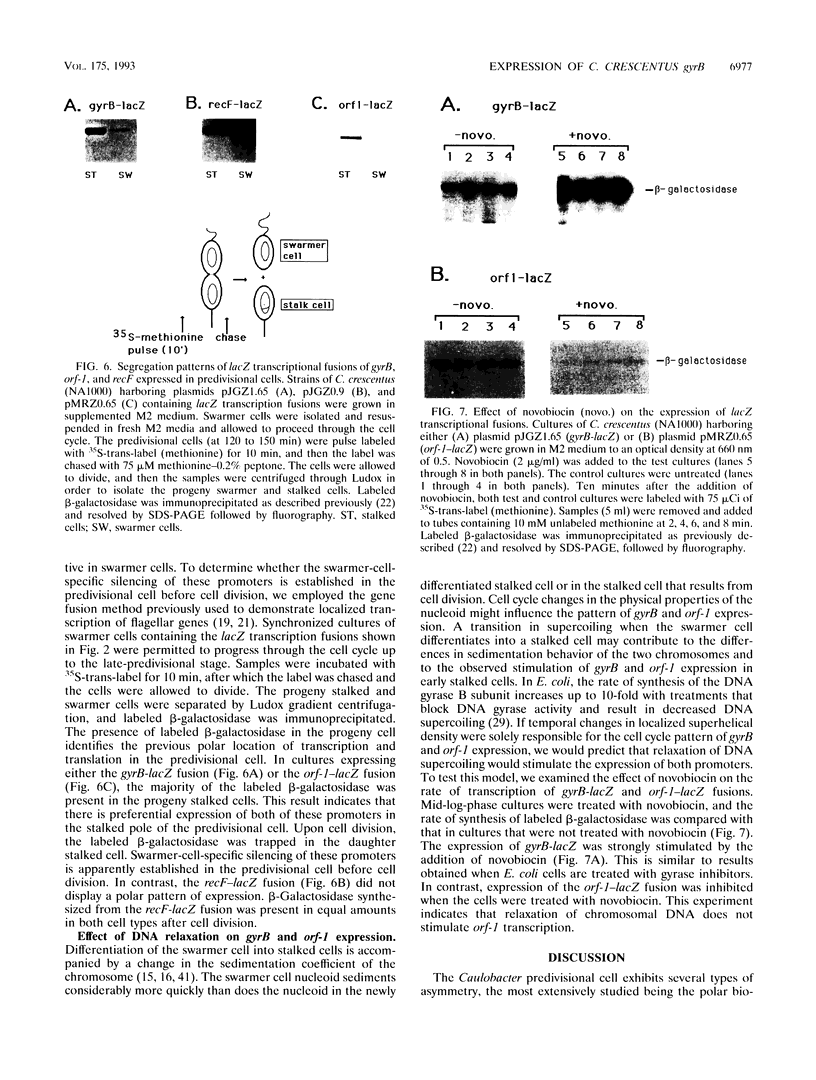
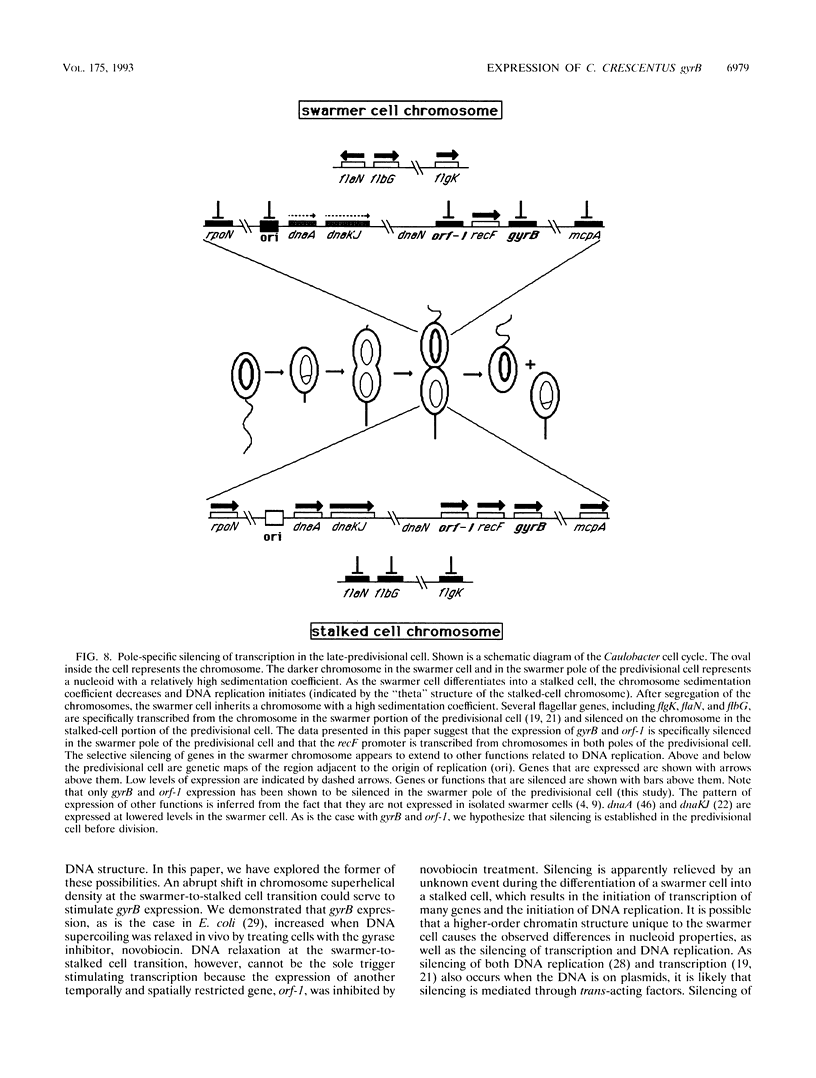
The bacterium Caulobacter crescentus undergoes an asymmetric cell division resulting in the formation of two different daughter cells, a motile swarmer cell and a nonmotile stalked cell. These two cell types differ in their program of gene expression, their ability to replicate DNA, and the physical properties of their nucleoids. We show here that two genes, gyrB (encoding the gyrase B subunit) and orf-1, are specifically transcribed from the chromosome in the portion of the predivisional cell destined for the progeny stalked cell. This is in contrast to a subset of flagellar genes which are transcribed from the chromosome in the incipient swarmer portion of the predivisional cell. gyrB and orf-1 are within a newly identified cluster of genes involved in DNA replication and recombination, including dnaN and recF. The transcription of gyrB and orf1 occurs from the replication-competent chromosome in stalked and predivisional cells and is silenced in swarmer cells. We hypothesize that selective silencing of groups of genes in the chromosomes at the swarmer and stalked poles of the predivisional cell results in the different developmental programs and the difference in replicative ability of the two progeny cells.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adachi T., Mizuuchi K., Menzel R., Gellert M. DNA sequence and transcription of the region upstream of the E. coli gyrB gene. Nucleic Acids Res. 1984 Aug 24;12(16):6389–6395. doi: 10.1093/nar/12.16.6389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adachi T., Mizuuchi M., Robinson E. A., Appella E., O'Dea M. H., Gellert M., Mizuuchi K. DNA sequence of the E. coli gyrB gene: application of a new sequencing strategy. Nucleic Acids Res. 1987 Jan 26;15(2):771–784. doi: 10.1093/nar/15.2.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali J. A., Jackson A. P., Howells A. J., Maxwell A. The 43-kilodalton N-terminal fragment of the DNA gyrase B protein hydrolyzes ATP and binds coumarin drugs. Biochemistry. 1993 Mar 16;32(10):2717–2724. doi: 10.1021/bi00061a033. [DOI] [PubMed] [Google Scholar]
- Alley M. R., Gomes S. L., Alexander W., Shapiro L. Genetic analysis of a temporally transcribed chemotaxis gene cluster in Caulobacter crescentus. Genetics. 1991 Oct;129(2):333–341. doi: 10.1093/genetics/129.2.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alley M. R., Maddock J. R., Shapiro L. Polar localization of a bacterial chemoreceptor. Genes Dev. 1992 May;6(5):825–836. doi: 10.1101/gad.6.5.825. [DOI] [PubMed] [Google Scholar]
- Alley M. R., Maddock J. R., Shapiro L. Requirement of the carboxyl terminus of a bacterial chemoreceptor for its targeted proteolysis. Science. 1993 Mar 19;259(5102):1754–1757. doi: 10.1126/science.8456303. [DOI] [PubMed] [Google Scholar]
- Amemiya K. Conserved sequence elements upstream and downstream from the transcription initiation site of the Caulobacter crescentus rrnA gene cluster. J Mol Biol. 1989 Nov 20;210(2):245–254. doi: 10.1016/0022-2836(89)90327-6. [DOI] [PubMed] [Google Scholar]
- Brun Y. V., Shapiro L. A temporally controlled sigma-factor is required for polar morphogenesis and normal cell division in Caulobacter. Genes Dev. 1992 Dec;6(12A):2395–2408. doi: 10.1101/gad.6.12a.2395. [DOI] [PubMed] [Google Scholar]
- Cheung K. K., Newton A. Patterns of protein synthesis during development in Caulobacter crescentus. Dev Biol. 1977 Apr;56(2):417–425. doi: 10.1016/0012-1606(77)90281-0. [DOI] [PubMed] [Google Scholar]
- Contreras I., Shapiro L., Henry S. Membrane phospholipid composition of Caulobacter crescentus. J Bacteriol. 1978 Sep;135(3):1130–1136. doi: 10.1128/jb.135.3.1130-1136.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dingwall A., Zhuang W. Y., Quon K., Shapiro L. Expression of an early gene in the flagellar regulatory hierarchy is sensitive to an interruption in DNA replication. J Bacteriol. 1992 Mar;174(6):1760–1768. doi: 10.1128/jb.174.6.1760-1768.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ditta G., Stanfield S., Corbin D., Helinski D. R. Broad host range DNA cloning system for gram-negative bacteria: construction of a gene bank of Rhizobium meliloti. Proc Natl Acad Sci U S A. 1980 Dec;77(12):7347–7351. doi: 10.1073/pnas.77.12.7347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drlica K., Worcel A. Conformational transitions in the Escherichia coli chromosome: analysis by viscometry and sedimentation. J Mol Biol. 1975 Oct 25;98(2):393–411. doi: 10.1016/s0022-2836(75)80126-4. [DOI] [PubMed] [Google Scholar]
- Ely B., Johnson R. C. Generalized Transduction in CAULOBACTER CRESCENTUS. Genetics. 1977 Nov;87(3):391–399. doi: 10.1093/genetics/87.3.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evinger M., Agabian N. Caulobacter crescentus nucleoid: analysis of sedimentation behavior and protein composition during the cell cycle. Proc Natl Acad Sci U S A. 1979 Jan;76(1):175–178. doi: 10.1073/pnas.76.1.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evinger M., Agabian N. Envelope-associated nucleoid from Caulobacter crescentus stalked and swarmer cells. J Bacteriol. 1977 Oct;132(1):294–301. doi: 10.1128/jb.132.1.294-301.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher J. A., Smit J., Agabian N. Transcriptional analysis of the major surface array gene of Caulobacter crescentus. J Bacteriol. 1988 Oct;170(10):4706–4713. doi: 10.1128/jb.170.10.4706-4713.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gober J. W., Alley M. R., Shapiro L. Positional information during Caulobacter cell differentiation. Curr Opin Genet Dev. 1991 Oct;1(3):324–329. doi: 10.1016/s0959-437x(05)80295-3. [DOI] [PubMed] [Google Scholar]
- Gober J. W., Champer R., Reuter S., Shapiro L. Expression of positional information during cell differentiation of Caulobacter. Cell. 1991 Jan 25;64(2):381–391. doi: 10.1016/0092-8674(91)90646-g. [DOI] [PubMed] [Google Scholar]
- Gober J. W., Shapiro L. A developmentally regulated Caulobacter flagellar promoter is activated by 3' enhancer and IHF binding elements. Mol Biol Cell. 1992 Aug;3(8):913–926. doi: 10.1091/mbc.3.8.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes S. L., Gober J. W., Shapiro L. Expression of the Caulobacter heat shock gene dnaK is developmentally controlled during growth at normal temperatures. J Bacteriol. 1990 Jun;172(6):3051–3059. doi: 10.1128/jb.172.6.3051-3059.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes S. L., Shapiro L. Differential expression and positioning of chemotaxis methylation proteins in Caulobacter. J Mol Biol. 1984 Sep 25;178(3):551–568. doi: 10.1016/0022-2836(84)90238-9. [DOI] [PubMed] [Google Scholar]
- Göransson M., Sondén B., Nilsson P., Dagberg B., Forsman K., Emanuelsson K., Uhlin B. E. Transcriptional silencing and thermoregulation of gene expression in Escherichia coli. Nature. 1990 Apr 12;344(6267):682–685. doi: 10.1038/344682a0. [DOI] [PubMed] [Google Scholar]
- Horvitz H. R., Herskowitz I. Mechanisms of asymmetric cell division: two Bs or not two Bs, that is the question. Cell. 1992 Jan 24;68(2):237–255. doi: 10.1016/0092-8674(92)90468-r. [DOI] [PubMed] [Google Scholar]
- Hromockyj A. E., Tucker S. C., Maurelli A. T. Temperature regulation of Shigella virulence: identification of the repressor gene virR, an analogue of hns, and partial complementation by tyrosyl transfer RNA (tRNA1(Tyr)). Mol Microbiol. 1992 Aug;6(15):2113–2124. doi: 10.1111/j.1365-2958.1992.tb01385.x. [DOI] [PubMed] [Google Scholar]
- Menzel R., Gellert M. Regulation of the genes for E. coli DNA gyrase: homeostatic control of DNA supercoiling. Cell. 1983 Aug;34(1):105–113. doi: 10.1016/0092-8674(83)90140-x. [DOI] [PubMed] [Google Scholar]
- Mizuuchi K., Mizuuchi M., O'Dea M. H., Gellert M. Cloning and simplified purification of Escherichia coli DNA gyrase A and B proteins. J Biol Chem. 1984 Jul 25;259(14):9199–9201. [PubMed] [Google Scholar]
- Newton A., Ohta N. Regulation of the cell division cycle and differentiation in bacteria. Annu Rev Microbiol. 1990;44:689–719. doi: 10.1146/annurev.mi.44.100190.003353. [DOI] [PubMed] [Google Scholar]
- Ohta N., Masurekar M., Newton A. Cloning and cell cycle-dependent expression of DNA replication gene dnaC from Caulobacter crescentus. J Bacteriol. 1990 Dec;172(12):7027–7034. doi: 10.1128/jb.172.12.7027-7034.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- POINDEXTER J. S. BIOLOGICAL PROPERTIES AND CLASSIFICATION OF THE CAULOBACTER GROUP. Bacteriol Rev. 1964 Sep;28:231–295. doi: 10.1128/br.28.3.231-295.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reece R. J., Maxwell A. DNA gyrase: structure and function. Crit Rev Biochem Mol Biol. 1991;26(3-4):335–375. doi: 10.3109/10409239109114072. [DOI] [PubMed] [Google Scholar]
- Reuter S. H., Shapiro L. Asymmetric segregation of heat-shock proteins upon cell division in Caulobacter crescentus. J Mol Biol. 1987 Apr 20;194(4):653–662. doi: 10.1016/0022-2836(87)90242-7. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spassky A., Rimsky S., Garreau H., Buc H. H1a, an E. coli DNA-binding protein which accumulates in stationary phase, strongly compacts DNA in vitro. Nucleic Acids Res. 1984 Jul 11;12(13):5321–5340. doi: 10.1093/nar/12.13.5321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Way S. M., Newton A., Mullin A. H., Mullin D. A. Identification of the promoter and a negative regulatory element, ftr4, that is needed for cell cycle timing of fliF operon expression in Caulobacter crescentus. J Bacteriol. 1993 Jan;175(2):367–376. doi: 10.1128/jb.175.2.367-376.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worcel A., Burgi E. On the structure of the folded chromosome of Escherichia coli. J Mol Biol. 1972 Nov 14;71(2):127–147. doi: 10.1016/0022-2836(72)90342-7. [DOI] [PubMed] [Google Scholar]
- Yu J., Shapiro L. Early Caulobacter crescentus genes fliL and fliM are required for flagellar gene expression and normal cell division. J Bacteriol. 1992 May;174(10):3327–3338. doi: 10.1128/jb.174.10.3327-3338.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]