Abstract
The major acidic exopolysaccharide of Rhizobium meliloti, termed succinoglycan, is required for nodule invasion and possibly nodule development. Succinoglycan is a polymer of octasaccharide subunits composed of one galactose residue, seven glucose residues, and acetyl, succinyl, and pyruvyl modifications, which is synthesized on an isoprenoid lipid carrier. A cluster of exo genes in R. meliloti are required for succinoglycan production, and the biosynthetic roles of their gene products have recently been determined (T.L. Reuber and G. C. Walker, Cell 74:269-280, 1993). Our sequencing of 16 kb of this cluster of exo genes and further genetic analysis of this region resulted in the discovery of several new exo genes and has allowed a correlation of the genetic map with the DNA sequence. In this paper we present the sequences of genes that are required for the addition of the succinyl and pyruvyl modifications to the lipid-linked intermediate and genes required for the polymerization of the octasaccharide subunits or the export of succinoglycan. In addition, on the basis of homologies to known proteins, we suggest that ExoN is a uridine diphosphoglucose pyrophosphorylase and that ExoK is a beta(1,3)-beta (1,4)-glucanase. We propose a model for succinoglycan biosynthesis and processing which assigns roles to the products of nineteen exo genes.
Full text
PDF








Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Battisti L., Lara J. C., Leigh J. A. Specific oligosaccharide form of the Rhizobium meliloti exopolysaccharide promotes nodule invasion in alfalfa. Proc Natl Acad Sci U S A. 1992 Jun 15;89(12):5625–5629. doi: 10.1073/pnas.89.12.5625. [DOI] [PMC free article] [PubMed] [Google Scholar]
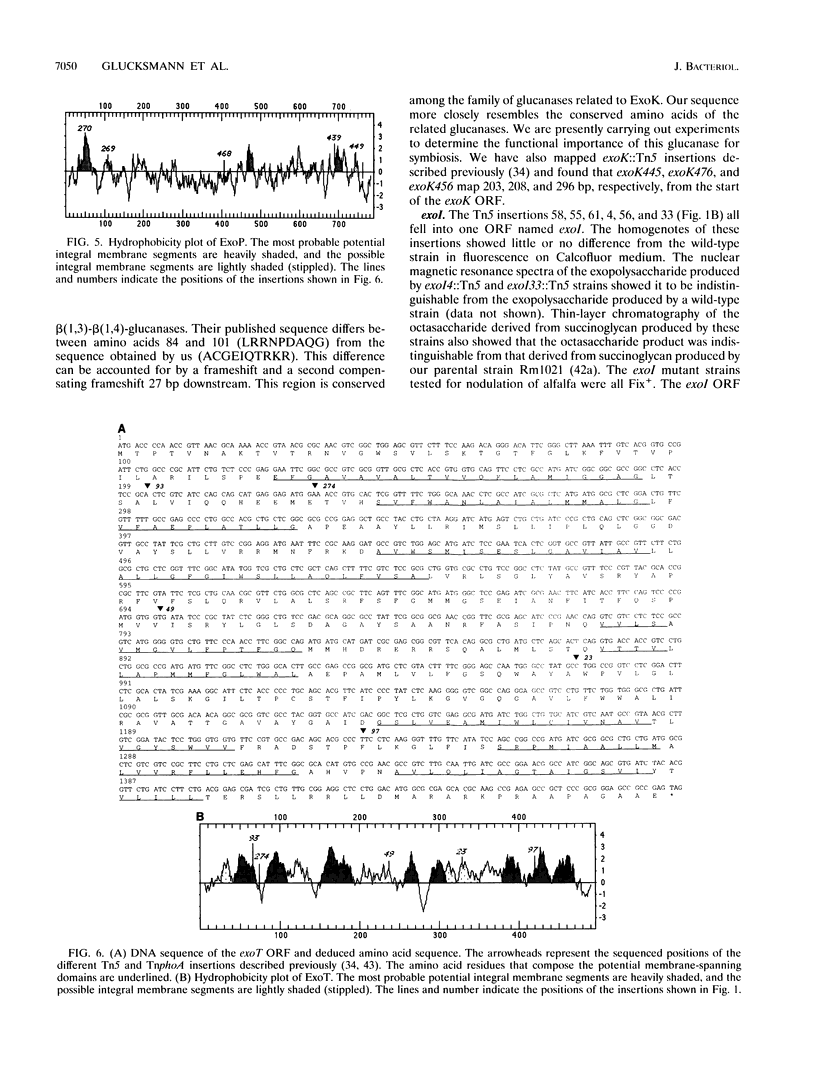
- Becker A., Kleickmann A., Arnold W., Pühler A. Analysis of the Rhizobium meliloti exoH/exoK/exoL fragment: ExoK shows homology to excreted endo-beta-1,3-1,4-glucanases and ExoH resembles membrane proteins. Mol Gen Genet. 1993 Apr;238(1-2):145–154. doi: 10.1007/BF00279541. [DOI] [PubMed] [Google Scholar]
- Boulnois G. J., Roberts I. S. Genetics of capsular polysaccharide production in bacteria. Curr Top Microbiol Immunol. 1990;150:1–18. doi: 10.1007/978-3-642-74694-9_1. [DOI] [PubMed] [Google Scholar]
- Brede G., Fjaervik E., Valla S. Nucleotide sequence and expression analysis of the Acetobacter xylinum uridine diphosphoglucose pyrophosphorylase gene. J Bacteriol. 1991 Nov;173(21):7042–7045. doi: 10.1128/jb.173.21.7042-7045.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buendia A. M., Enenkel B., Köplin R., Niehaus K., Arnold W., Pühler A. The Rhizobium meliloti exoZl exoB fragment of megaplasmid 2: ExoB functions as a UDP-glucose 4-epimerase and ExoZ shows homology to NodX of Rhizobium leguminosarum biovar viciae strain TOM. Mol Microbiol. 1991 Jun;5(6):1519–1530. doi: 10.1111/j.1365-2958.1991.tb00799.x. [DOI] [PubMed] [Google Scholar]
- Canter Cremers H. C., Batley M., Redmond J. W., Eydems L., Breedveld M. W., Zevehuizen L. P., Pees E., Wijffelman C. A., Lugtenberg B. J. Rhizobium leguminosarum exoB mutants are deficient in the synthesis of UDP-glucose 4'-epimerase. J Biol Chem. 1990 Dec 5;265(34):21122–21127. [PubMed] [Google Scholar]
- Distler J., Ebert A., Mansouri K., Pissowotzki K., Stockmann M., Piepersberg W. Gene cluster for streptomycin biosynthesis in Streptomyces griseus: nucleotide sequence of three genes and analysis of transcriptional activity. Nucleic Acids Res. 1987 Oct 12;15(19):8041–8056. doi: 10.1093/nar/15.19.8041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ditta G., Schmidhauser T., Yakobson E., Lu P., Liang X. W., Finlay D. R., Guiney D., Helinski D. R. Plasmids related to the broad host range vector, pRK290, useful for gene cloning and for monitoring gene expression. Plasmid. 1985 Mar;13(2):149–153. doi: 10.1016/0147-619x(85)90068-x. [DOI] [PubMed] [Google Scholar]
- Finan T. M., Hirsch A. M., Leigh J. A., Johansen E., Kuldau G. A., Deegan S., Walker G. C., Signer E. R. Symbiotic mutants of Rhizobium meliloti that uncouple plant from bacterial differentiation. Cell. 1985 Apr;40(4):869–877. doi: 10.1016/0092-8674(85)90346-0. [DOI] [PubMed] [Google Scholar]
- Finan T. M., Kunkel B., De Vos G. F., Signer E. R. Second symbiotic megaplasmid in Rhizobium meliloti carrying exopolysaccharide and thiamine synthesis genes. J Bacteriol. 1986 Jul;167(1):66–72. doi: 10.1128/jb.167.1.66-72.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glucksmann M. A., Reuber T. L., Walker G. C. Family of glycosyl transferases needed for the synthesis of succinoglycan by Rhizobium meliloti. J Bacteriol. 1993 Nov;175(21):7033–7044. doi: 10.1128/jb.175.21.7033-7044.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray J. X., Djordjevic M. A., Rolfe B. G. Two genes that regulate exopolysaccharide production in Rhizobium sp. strain NGR234: DNA sequences and resultant phenotypes. J Bacteriol. 1990 Jan;172(1):193–203. doi: 10.1128/jb.172.1.193-203.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins C. F. ABC transporters: from microorganisms to man. Annu Rev Cell Biol. 1992;8:67–113. doi: 10.1146/annurev.cb.08.110192.000435. [DOI] [PubMed] [Google Scholar]
- Jiang X. M., Neal B., Santiago F., Lee S. J., Romana L. K., Reeves P. R. Structure and sequence of the rfb (O antigen) gene cluster of Salmonella serovar typhimurium (strain LT2). Mol Microbiol. 1991 Mar;5(3):695–713. doi: 10.1111/j.1365-2958.1991.tb00741.x. [DOI] [PubMed] [Google Scholar]
- Klein P., Kanehisa M., DeLisi C. The detection and classification of membrane-spanning proteins. Biochim Biophys Acta. 1985 May 28;815(3):468–476. doi: 10.1016/0005-2736(85)90375-x. [DOI] [PubMed] [Google Scholar]
- Kyte J., Doolittle R. F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982 May 5;157(1):105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Leigh J. A., Coplin D. L. Exopolysaccharides in plant-bacterial interactions. Annu Rev Microbiol. 1992;46:307–346. doi: 10.1146/annurev.mi.46.100192.001515. [DOI] [PubMed] [Google Scholar]
- Leigh J. A., Lee C. C. Characterization of polysaccharides of Rhizobium meliloti exo mutants that form ineffective nodules. J Bacteriol. 1988 Aug;170(8):3327–3332. doi: 10.1128/jb.170.8.3327-3332.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leigh J. A., Reed J. W., Hanks J. F., Hirsch A. M., Walker G. C. Rhizobium meliloti mutants that fail to succinylate their calcofluor-binding exopolysaccharide are defective in nodule invasion. Cell. 1987 Nov 20;51(4):579–587. doi: 10.1016/0092-8674(87)90127-9. [DOI] [PubMed] [Google Scholar]
- Leigh J. A., Signer E. R., Walker G. C. Exopolysaccharide-deficient mutants of Rhizobium meliloti that form ineffective nodules. Proc Natl Acad Sci U S A. 1985 Sep;82(18):6231–6235. doi: 10.1073/pnas.82.18.6231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lever M. A new reaction for colorimetric determination of carbohydrates. Anal Biochem. 1972 May;47(1):273–279. doi: 10.1016/0003-2697(72)90301-6. [DOI] [PubMed] [Google Scholar]
- Lipman D. J., Pearson W. R. Rapid and sensitive protein similarity searches. Science. 1985 Mar 22;227(4693):1435–1441. doi: 10.1126/science.2983426. [DOI] [PubMed] [Google Scholar]
- Long S., McCune S., Walker G. C. Symbiotic loci of Rhizobium meliloti identified by random TnphoA mutagenesis. J Bacteriol. 1988 Sep;170(9):4257–4265. doi: 10.1128/jb.170.9.4257-4265.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long S., Reed J. W., Himawan J., Walker G. C. Genetic analysis of a cluster of genes required for synthesis of the calcofluor-binding exopolysaccharide of Rhizobium meliloti. J Bacteriol. 1988 Sep;170(9):4239–4248. doi: 10.1128/jb.170.9.4239-4248.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MELO A., GLASER L. THE NUCLEOTIDE SPECIFICITY AND FEEDBACK CONTROL OF THYMIDINE DIPHOSPHATE D-GLUCOSE PYROPHOSPHORYLASE. J Biol Chem. 1965 Jan;240:398–405. [PubMed] [Google Scholar]
- Marino P. A., Phan K. A., Osborn M. J. Energy dependence of lipopolysaccharide translocation in Salmonella typhimurium. J Biol Chem. 1985 Dec 5;260(28):14965–14970. [PubMed] [Google Scholar]
- Müller P., Keller M., Weng W. M., Quandt J., Arnold W., Pühler A. Genetic analysis of the Rhizobium meliloti exoYFQ operon: ExoY is homologous to sugar transferases and ExoQ represents a transmembrane protein. Mol Plant Microbe Interact. 1993 Jan-Feb;6(1):55–65. doi: 10.1094/mpmi-6-055. [DOI] [PubMed] [Google Scholar]
- Nakae T., Nikaido H. Multiple molecular forms of uridine diphosphate glucose pyrophosphorylase from Salmonella typhimurium. I. Catalytic properties of various forms. J Biol Chem. 1971 Jul 25;246(14):4386–4396. [PubMed] [Google Scholar]
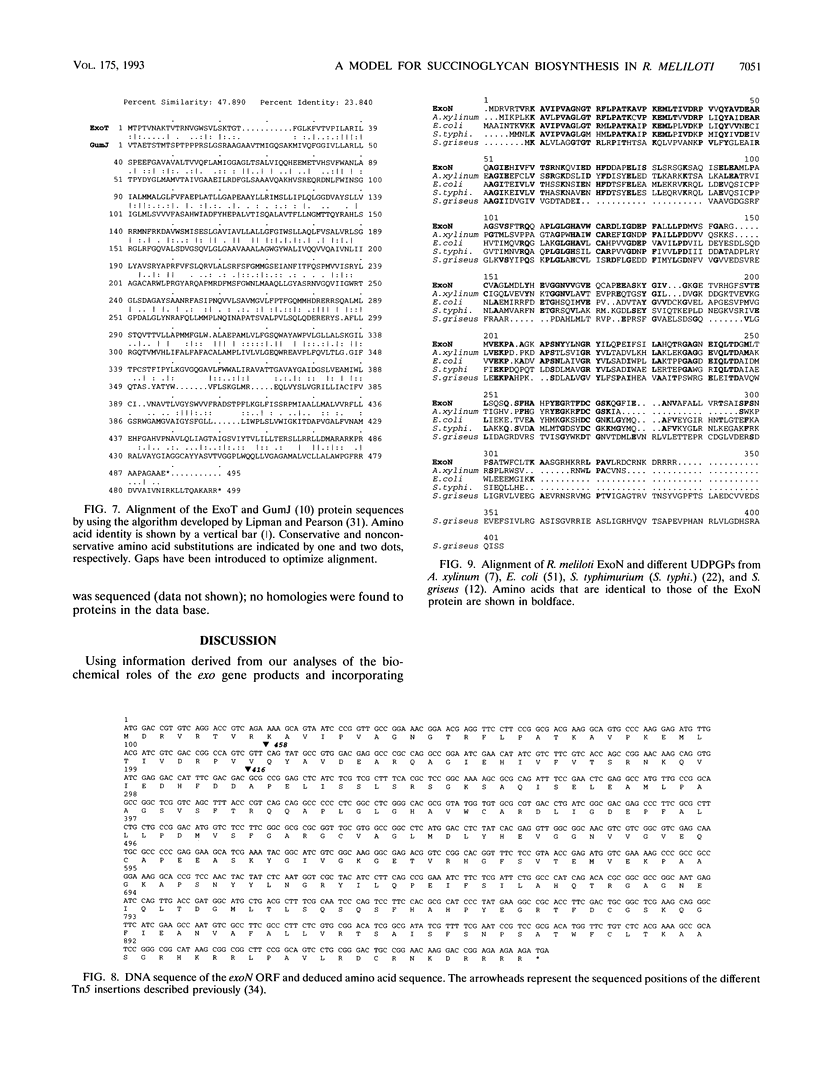
- Reed J. W., Capage M., Walker G. C. Rhizobium meliloti exoG and exoJ mutations affect the exoX-exoY system for modulation of exopolysaccharide production. J Bacteriol. 1991 Jun;173(12):3776–3788. doi: 10.1128/jb.173.12.3776-3788.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuber T. L., Long S., Walker G. C. Regulation of Rhizobium meliloti exo genes in free-living cells and in planta examined by using TnphoA fusions. J Bacteriol. 1991 Jan;173(2):426–434. doi: 10.1128/jb.173.2.426-434.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuber T. L., Walker G. C. Biosynthesis of succinoglycan, a symbiotically important exopolysaccharide of Rhizobium meliloti. Cell. 1993 Jul 30;74(2):269–280. doi: 10.1016/0092-8674(93)90418-p. [DOI] [PubMed] [Google Scholar]
- Reuber T. L., Walker G. C. The acetyl substituent of succinoglycan is not necessary for alfalfa nodule invasion by Rhizobium meliloti Rm1021. J Bacteriol. 1993 Jun;175(11):3653–3655. doi: 10.1128/jb.175.11.3653-3655.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruvkun G. B., Sundaresan V., Ausubel F. M. Directed transposon Tn5 mutagenesis and complementation analysis of Rhizobium meliloti symbiotic nitrogen fixation genes. Cell. 1982 Jun;29(2):551–559. doi: 10.1016/0092-8674(82)90171-4. [DOI] [PubMed] [Google Scholar]
- Shibaev V. N. Biosynthesis of bacterial polysaccharide chains composed of repeating units. Adv Carbohydr Chem Biochem. 1986;44:277–339. doi: 10.1016/s0065-2318(08)60080-3. [DOI] [PubMed] [Google Scholar]
- Silver R. P., Vann W. F., Aaronson W. Genetic and molecular analyses of Escherichia coli K1 antigen genes. J Bacteriol. 1984 Feb;157(2):568–575. doi: 10.1128/jb.157.2.568-575.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith A. N., Boulnois G. J., Roberts I. S. Molecular analysis of the Escherichia coli K5 kps locus: identification and characterization of an inner-membrane capsular polysaccharide transport system. Mol Microbiol. 1990 Nov;4(11):1863–1869. doi: 10.1111/j.1365-2958.1990.tb02035.x. [DOI] [PubMed] [Google Scholar]
- Stanfield S. W., Ielpi L., O'Brochta D., Helinski D. R., Ditta G. S. The ndvA gene product of Rhizobium meliloti is required for beta-(1----2)glucan production and has homology to the ATP-binding export protein HlyB. J Bacteriol. 1988 Aug;170(8):3523–3530. doi: 10.1128/jb.170.8.3523-3530.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueguchi C., Ito K. Multicopy suppression: an approach to understanding intracellular functioning of the protein export system. J Bacteriol. 1992 Mar;174(5):1454–1461. doi: 10.1128/jb.174.5.1454-1461.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urzainqui A., Walker G. C. Exogenous suppression of the symbiotic deficiencies of Rhizobium meliloti exo mutants. J Bacteriol. 1992 May;174(10):3403–3406. doi: 10.1128/jb.174.10.3403-3406.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uttaro A. D., Cangelosi G. A., Geremia R. A., Nester E. W., Ugalde R. A. Biochemical characterization of avirulent exoC mutants of Agrobacterium tumefaciens. J Bacteriol. 1990 Mar;172(3):1640–1646. doi: 10.1128/jb.172.3.1640-1646.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood D., Darlison M. G., Wilde R. J., Guest J. R. Nucleotide sequence encoding the flavoprotein and hydrophobic subunits of the succinate dehydrogenase of Escherichia coli. Biochem J. 1984 Sep 1;222(2):519–534. doi: 10.1042/bj2220519. [DOI] [PMC free article] [PubMed] [Google Scholar]


