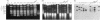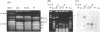Abstract
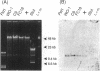
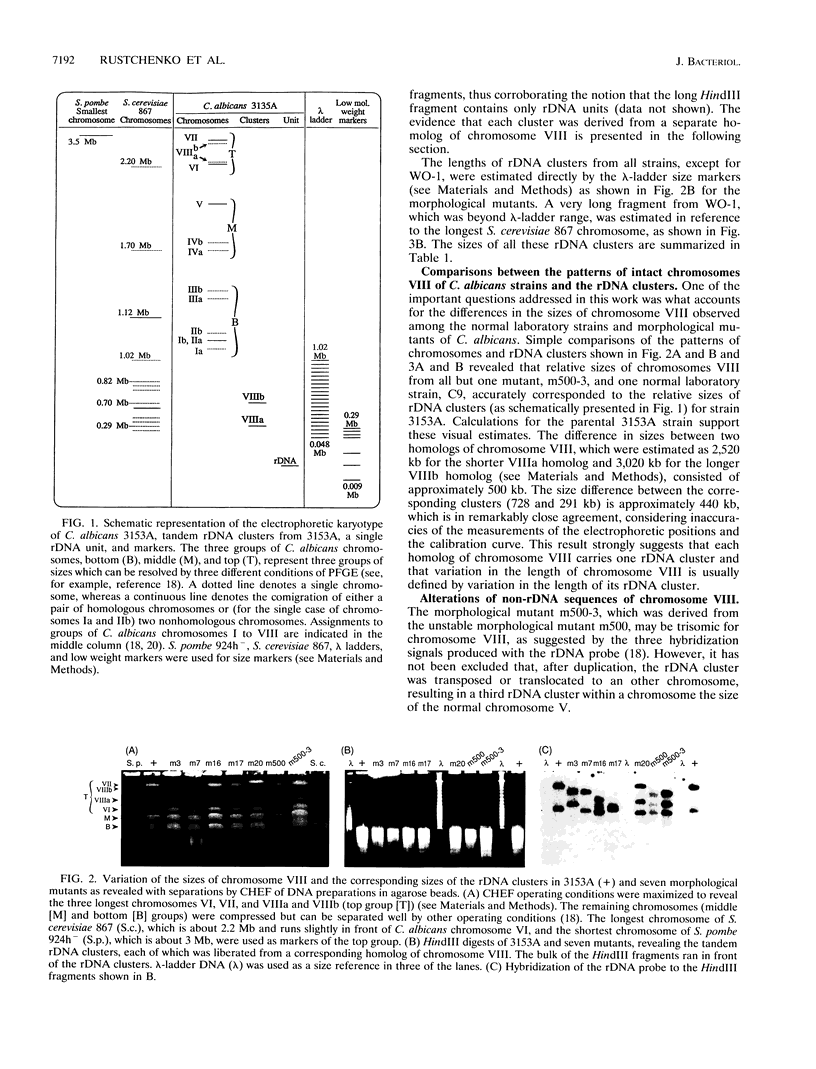
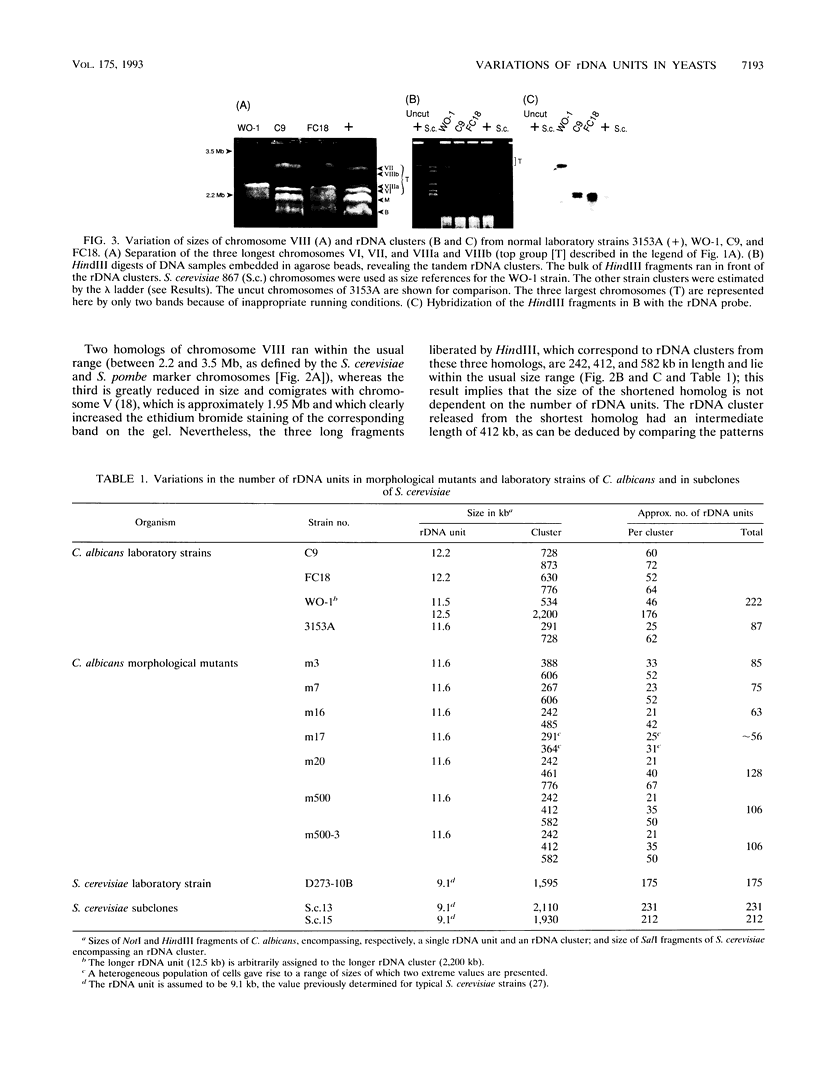
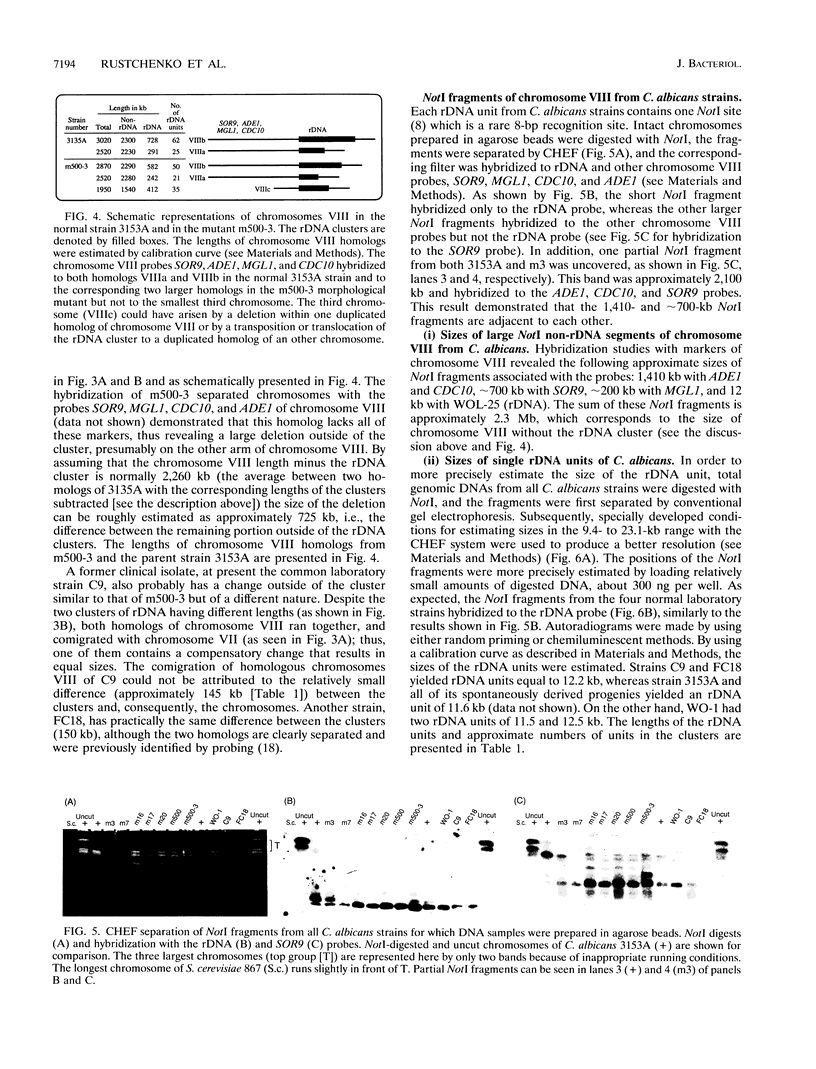
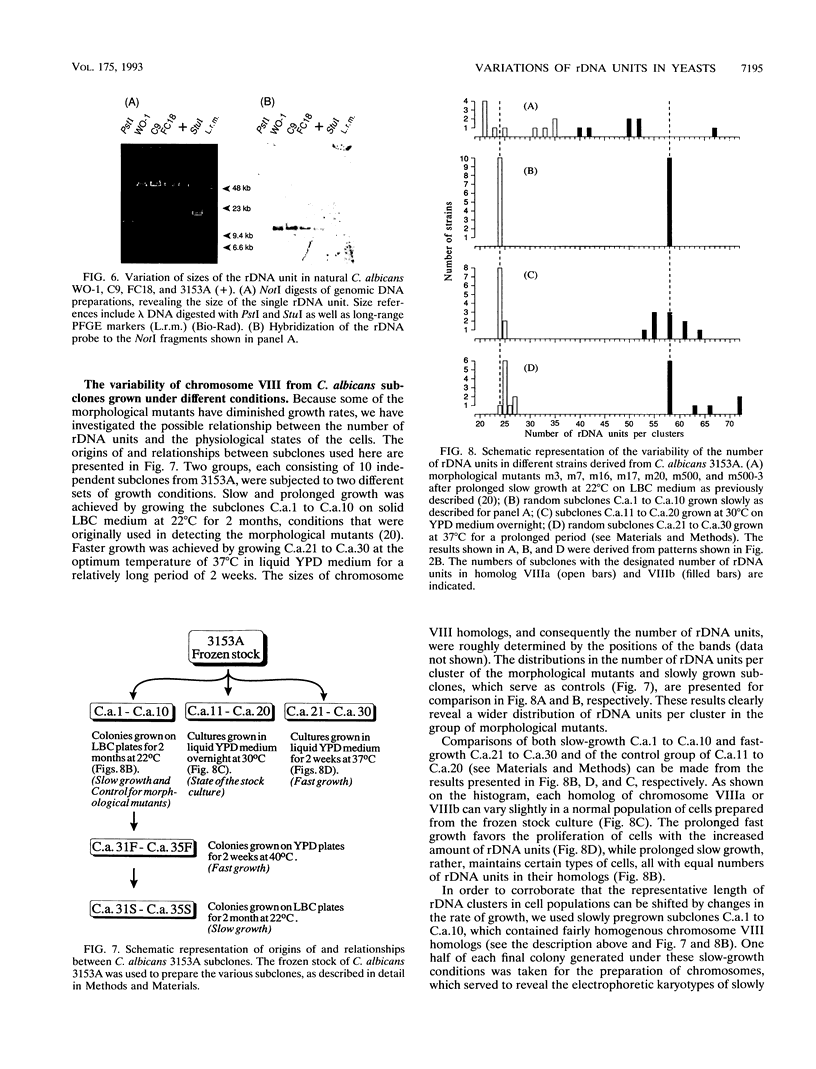
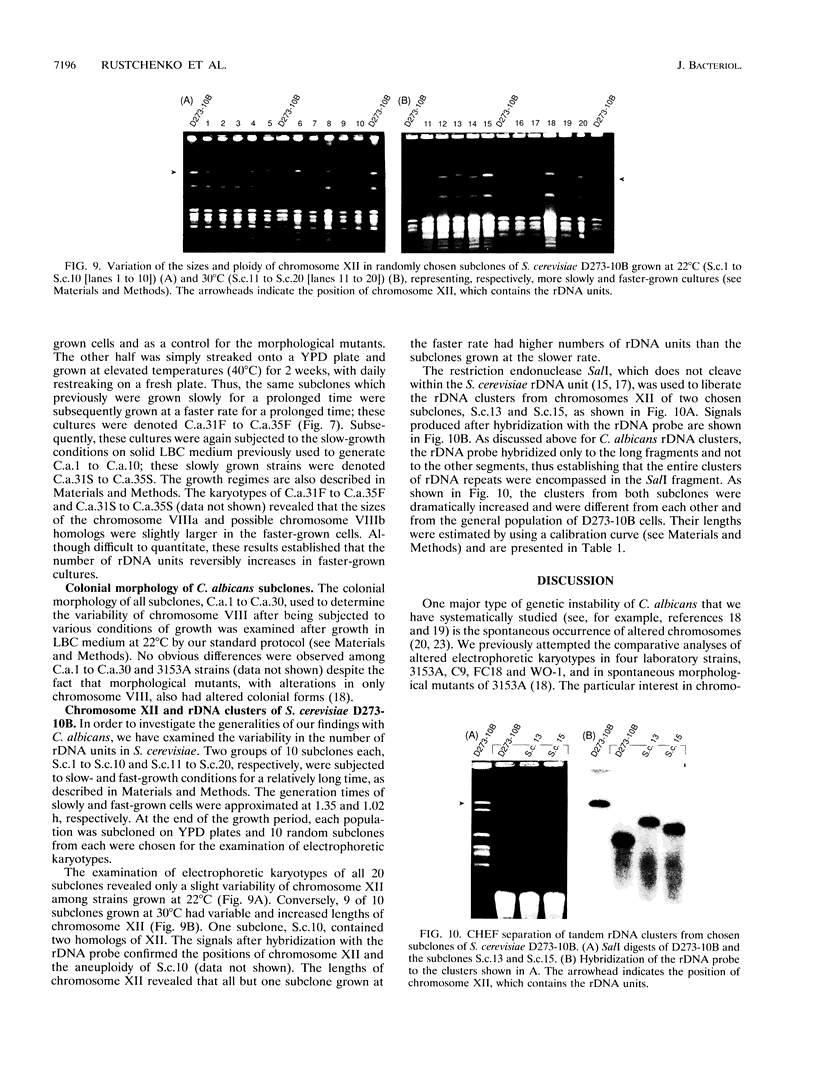
Naturally occurring strains of Candida albicans are opportunistic pathogens that lack a sexual cycle and that are usually diploids with eight pairs of chromosomes. C. albicans spontaneously gives rise to a high frequency of colonial morphology mutants with altered electrophoretic karyotypes, involving one or more of their chromosomes. However, the most frequent changes involve chromosome VIII, which contains the genes coding for ribosomal DNA (rDNA) units. We have used restriction fragment lengths to analyze the number and physical array of the rDNA units on chromosome VIII in four normal clinical strains and seven morphological mutants derived spontaneously from one of the clinical isolates. HindIII does not cleave the rDNA repeats and liberates the tandem rDNA cluster from each homolog of chromosome VIII as a single fragment, whereas the cleavage at a single site by NotI reveals the size of the single rDNA unit. All clinical strains and morphological mutants differed greatly in the number of rDNA units per cluster and per cell. The four clinical isolates differed additionally among themselves by the size of the single rDNA unit. For a total of 25 chromosome VIII homologs in a total of 11 strains considered, the variability of chromosome VIII was exclusively due to the length of rDNA clusters (or the number of rDNA units) in approximately 92% of the cases, whereas the others involved other rearrangements of chromosome VIII. Only slight variations in the number of rDNA units were observed among 10 random C. albicans subclones and 10 random Saccharomyces cerevisiae subclones grown for a prolonged time at 22 degrees C. However, when grown faster at optimal temperatures of 37 and 30 degrees C, respectively, both fungi accumulated higher numbers of rDNA units, suggesting that this condition is selected for in rapidly growing cells. The morphological mutants, in comparison with the C. albicans subclones, contained a markedly wider distribution of the number of rDNA units, suggesting that a distinct process may be involved in altering the number of rDNA units in these mutants.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Asakura K., Iwaguchi S., Homma M., Sukai T., Higashide K., Tanaka K. Electrophoretic karyotypes of clinically isolated yeasts of Candida albicans and C. glabrata. J Gen Microbiol. 1991 Nov;137(11):2531–2538. doi: 10.1099/00221287-137-11-2531. [DOI] [PubMed] [Google Scholar]
- Carle G. F., Olson M. V. An electrophoretic karyotype for yeast. Proc Natl Acad Sci U S A. 1985 Jun;82(11):3756–3760. doi: 10.1073/pnas.82.11.3756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu G., Vollrath D., Davis R. W. Separation of large DNA molecules by contour-clamped homogeneous electric fields. Science. 1986 Dec 19;234(4783):1582–1585. doi: 10.1126/science.3538420. [DOI] [PubMed] [Google Scholar]
- Fan J. B., Chikashige Y., Smith C. L., Niwa O., Yanagida M., Cantor C. R. Construction of a Not I restriction map of the fission yeast Schizosaccharomyces pombe genome. Nucleic Acids Res. 1989 Apr 11;17(7):2801–2818. doi: 10.1093/nar/17.7.2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983 Jul 1;132(1):6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Iwaguchi S., Homma M., Tanaka K. Clonal variation of chromosome size derived from the rDNA cluster region in Candida albicans. J Gen Microbiol. 1992 Jun;138(6):1177–1184. doi: 10.1099/00221287-138-6-1177. [DOI] [PubMed] [Google Scholar]
- Iwaguchi S., Homma M., Tanaka K. Variation in the electrophoretic karyotype analysed by the assignment of DNA probes in Candida albicans. J Gen Microbiol. 1990 Dec;136(12):2433–2442. doi: 10.1099/00221287-136-12-2433. [DOI] [PubMed] [Google Scholar]
- Jakubczak J. L., Xiong Y., Eickbush T. H. Type I (R1) and type II (R2) ribosomal DNA insertions of Drosophila melanogaster are retrotransposable elements closely related to those of Bombyx mori. J Mol Biol. 1990 Mar 5;212(1):37–52. doi: 10.1016/0022-2836(90)90303-4. [DOI] [PubMed] [Google Scholar]
- Jemtland R., Maehlum E., Gabrielsen O. S., Oyen T. B. Regular distribution of length heterogeneities within non-transcribed spacer regions of cloned and genomic rDNA of Saccharomyces cerevisiae. Nucleic Acids Res. 1986 Jul 11;14(13):5145–5158. doi: 10.1093/nar/14.13.5145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasker B. A., Carle G. F., Kobayashi G. S., Medoff G. Comparison of the separation of Candida albicans chromosome-sized DNA by pulsed-field gel electrophoresis techniques. Nucleic Acids Res. 1989 May 25;17(10):3783–3793. doi: 10.1093/nar/17.10.3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K. L., Buckley H. R., Campbell C. C. An amino acid liquid synthetic medium for the development of mycelial and yeast forms of Candida Albicans. Sabouraudia. 1975 Jul;13(2):148–153. doi: 10.1080/00362177585190271. [DOI] [PubMed] [Google Scholar]
- Lott T. J., Boiron P., Reiss E. An electrophoretic karyotype for Candida albicans reveals large chromosomes in multiples. Mol Gen Genet. 1987 Aug;209(1):170–174. doi: 10.1007/BF00329854. [DOI] [PubMed] [Google Scholar]
- Merz W. G., Connelly C., Hieter P. Variation of electrophoretic karyotypes among clinical isolates of Candida albicans. J Clin Microbiol. 1988 May;26(5):842–845. doi: 10.1128/jcm.26.5.842-845.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasero P., Marilley M. Size variation of rDNA clusters in the yeasts Saccharomyces cerevisiae and Schizosaccharomyces pombe. Mol Gen Genet. 1993 Jan;236(2-3):448–452. doi: 10.1007/BF00277147. [DOI] [PubMed] [Google Scholar]
- Rustchenko-Bulgac E. P., Howard D. H. Multiple chromosomal and phenotypic changes in spontaneous mutants of Candida albicans. J Gen Microbiol. 1993 Jun;139(Pt 6):1195–1207. doi: 10.1099/00221287-139-6-1195. [DOI] [PubMed] [Google Scholar]
- Rustchenko-Bulgac E. P., Sherman F., Hicks J. B. Chromosomal rearrangements associated with morphological mutants provide a means for genetic variation of Candida albicans. J Bacteriol. 1990 Mar;172(3):1276–1283. doi: 10.1128/jb.172.3.1276-1283.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rustchenko-Bulgac E. P. Variations of Candida albicans electrophoretic karyotypes. J Bacteriol. 1991 Oct;173(20):6586–6596. doi: 10.1128/jb.173.20.6586-6596.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F. Getting started with yeast. Methods Enzymol. 1991;194:3–21. doi: 10.1016/0076-6879(91)94004-v. [DOI] [PubMed] [Google Scholar]
- Suzuki T., Kobayashi I., Kanbe T., Tanaka K. High frequency variation of colony morphology and chromosome reorganization in the pathogenic yeast Candida albicans. J Gen Microbiol. 1989 Feb;135(Pt 2):425–434. doi: 10.1099/00221287-135-2-425. [DOI] [PubMed] [Google Scholar]
- Szostak J. W., Wu R. Unequal crossing over in the ribosomal DNA of Saccharomyces cerevisiae. Nature. 1980 Apr 3;284(5755):426–430. doi: 10.1038/284426a0. [DOI] [PubMed] [Google Scholar]
- Wakem L. P., Sherman F. Chromosomal assignment of mutations by specific chromosome loss in the yeast Saccharomyces cerevisiae. Genetics. 1990 Jun;125(2):333–340. doi: 10.1093/genetics/125.2.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickes B., Staudinger J., Magee B. B., Kwon-Chung K. J., Magee P. T., Scherer S. Physical and genetic mapping of Candida albicans: several genes previously assigned to chromosome 1 map to chromosome R, the rDNA-containing linkage group. Infect Immun. 1991 Jul;59(7):2480–2484. doi: 10.1128/iai.59.7.2480-2484.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y., Eickbush T. H. The site-specific ribosomal DNA insertion element R1Bm belongs to a class of non-long-terminal-repeat retrotransposons. Mol Cell Biol. 1988 Jan;8(1):114–123. doi: 10.1128/mcb.8.1.114. [DOI] [PMC free article] [PubMed] [Google Scholar]