Abstract
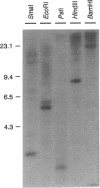
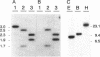
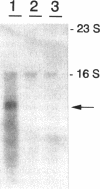
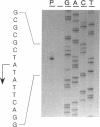
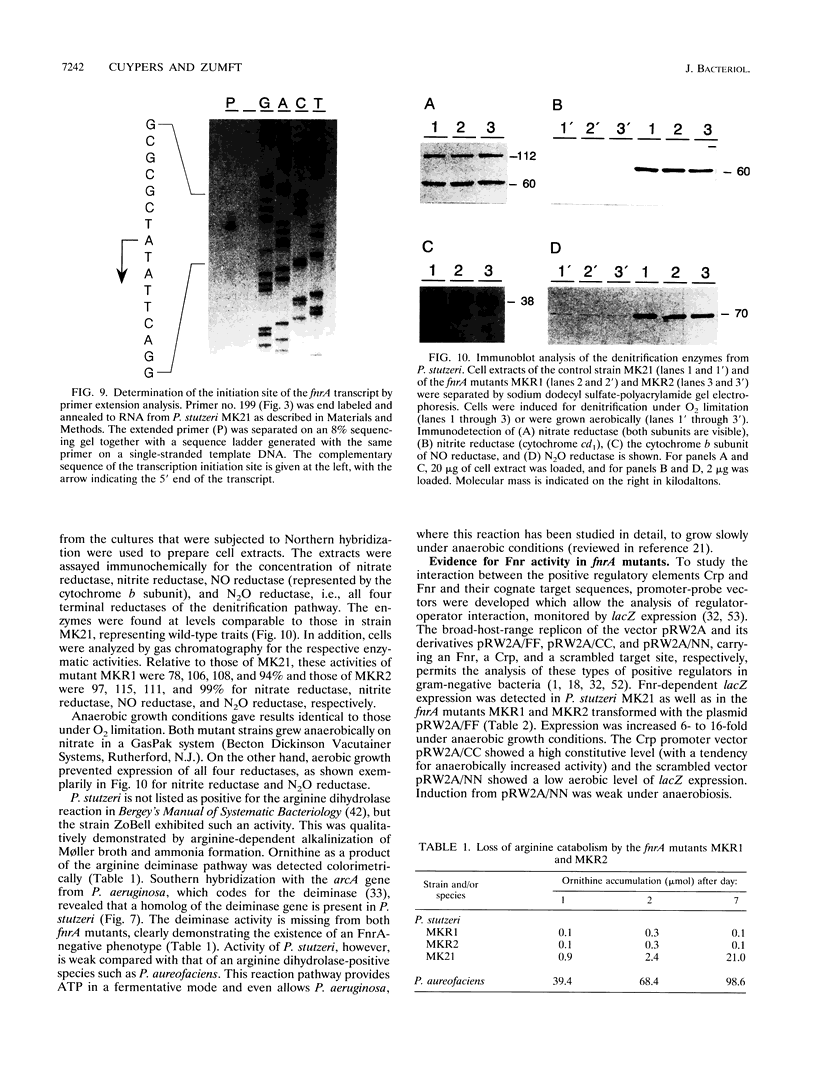
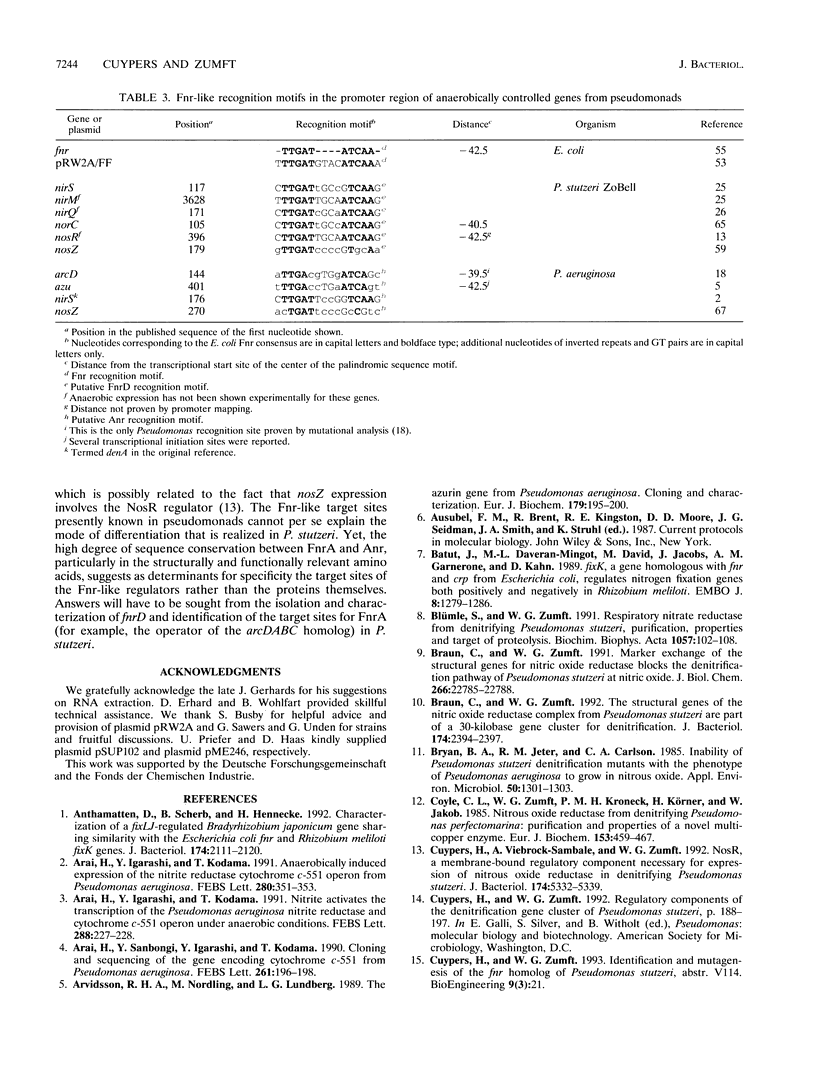
The synthesis of proteins necessary for the respiratory reduction of nitrate to dinitrogen is induced in most denitrifying bacteria by a shift to anaerobiosis. A homolog of the fur gene, which encodes a redox-active transcriptional activator in Escherichia coli, was isolated from Pseudomonas stutzeri by using the anr gene of Pseudomonas aeruginosa as the hybridization probe (R. G. Sawers, Mol. Microbiol. 5:1469-1481, 1991). The coding region was located on a 3-kb SmaI fragment. An open reading frame of 735 nucleotides, designated fnrA, had the coding potential for a protein of 244 amino acids (M(r) = 27,089) with 51.2% positional identity to the Fnr protein of E. coli and 86.1% to the Anr protein of P. aeruginosa. The fnrA gene gave a single transcript of 0.85 kb and complemented nitrate-dependent anaerobic growth of an fnr deletion mutant of E. coli. An open reading frame immediately downstream of fnrA encoded adenine phosphoribosyltransferase (EC 2.4.2.7). Mutations in fnrA were generated in vitro by insertional mutagenesis followed by gene replacement. Gene inactivation was shown by loss of the fnrA transcript and detection of an arginine deiminase (EC 3.5.3.6)-negative phenotype in the mutants. However, neither the enzymatic activities nor the levels of anaerobic expression of the respiratory enzymes nitrate reductase (EC 1.7.99.4), nitrate reductase (EC 1.9.3.2), NO reductase (EC 1.7.99.7), and N2O reductase (EC 1.7.99.6) were changed in fnrA mutants versus the P. stutzeri wild type. A promoter-probe vector for Fnr-dependent transcription was activated anaerobically in the fnrA mutants, suggesting the existence of a second Fnr homolog in the same bacterium. The Fnr-binding motifs, apparent in the promoter region of genes encoding denitrification components of P. stutzeri, are likely to be recognized by this second Fnr homolog. Preliminary evidence indicates also the presence of the catabolite activator protein, Crp, in P. stutzeri.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anthamatten D., Scherb B., Hennecke H. Characterization of a fixLJ-regulated Bradyrhizobium japonicum gene sharing similarity with the Escherichia coli fnr and Rhizobium meliloti fixK genes. J Bacteriol. 1992 Apr;174(7):2111–2120. doi: 10.1128/jb.174.7.2111-2120.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arai H., Igarashi Y., Kodama T. Anaerobically induced expression of the nitrite reductase cytochrome c-551 operon from Pseudomonas aeruginosa. FEBS Lett. 1991 Mar 25;280(2):351–353. doi: 10.1016/0014-5793(91)80329-2. [DOI] [PubMed] [Google Scholar]
- Arai H., Igarashi Y., Kodama T. Nitrite activates the transcription of the Pseudomonas aeruginosa nitrite reductase and cytochrome c-551 operon under anaerobic conditions. FEBS Lett. 1991 Aug 19;288(1-2):227–228. doi: 10.1016/0014-5793(91)81040-f. [DOI] [PubMed] [Google Scholar]
- Arai H., Sanbongi Y., Igarashi Y., Kodama T. Cloning and sequencing of the gene encoding cytochrome c-551 from Pseudomonas aeruginosa. FEBS Lett. 1990 Feb 12;261(1):196–198. doi: 10.1016/0014-5793(90)80669-a. [DOI] [PubMed] [Google Scholar]
- Arvidsson R. H., Nordling M., Lundberg L. G. The azurin gene from Pseudomonas aeruginosa. Cloning and characterization. Eur J Biochem. 1989 Jan 15;179(1):195–200. doi: 10.1111/j.1432-1033.1989.tb14540.x. [DOI] [PubMed] [Google Scholar]
- Batut J., Daveran-Mingot M. L., David M., Jacobs J., Garnerone A. M., Kahn D. fixK, a gene homologous with fnr and crp from Escherichia coli, regulates nitrogen fixation genes both positively and negatively in Rhizobium meliloti. EMBO J. 1989 Apr;8(4):1279–1286. doi: 10.1002/j.1460-2075.1989.tb03502.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun C., Zumft W. G. Marker exchange of the structural genes for nitric oxide reductase blocks the denitrification pathway of Pseudomonas stutzeri at nitric oxide. J Biol Chem. 1991 Dec 5;266(34):22785–22788. [PubMed] [Google Scholar]
- Braun C., Zumft W. G. The structural genes of the nitric oxide reductase complex from Pseudomonas stutzeri are part of a 30-kilobase gene cluster for denitrification. J Bacteriol. 1992 Apr;174(7):2394–2397. doi: 10.1128/jb.174.7.2394-2397.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryan B. A., Jeter R. M., Carlson C. A. Inability of Pseudomonas stutzeri denitrification mutants with the phenotype of Pseudomonas aeruginosa to grow in nitrous oxide. Appl Environ Microbiol. 1985 Nov;50(5):1301–1303. doi: 10.1128/aem.50.5.1301-1303.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyle C. L., Zumft W. G., Kroneck P. M., Körner H., Jakob W. Nitrous oxide reductase from denitrifying Pseudomonas perfectomarina. Purification and properties of a novel multicopper enzyme. Eur J Biochem. 1985 Dec 16;153(3):459–467. doi: 10.1111/j.1432-1033.1985.tb09324.x. [DOI] [PubMed] [Google Scholar]
- Cuypers H., Viebrock-Sambale A., Zumft W. G. NosR, a membrane-bound regulatory component necessary for expression of nitrous oxide reductase in denitrifying Pseudomonas stutzeri. J Bacteriol. 1992 Aug;174(16):5332–5339. doi: 10.1128/jb.174.16.5332-5339.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dispensa M., Thomas C. T., Kim M. K., Perrotta J. A., Gibson J., Harwood C. S. Anaerobic growth of Rhodopseudomonas palustris on 4-hydroxybenzoate is dependent on AadR, a member of the cyclic AMP receptor protein family of transcriptional regulators. J Bacteriol. 1992 Sep;174(18):5803–5813. doi: 10.1128/jb.174.18.5803-5813.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983 Jul 1;132(1):6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Galimand M., Gamper M., Zimmermann A., Haas D. Positive FNR-like control of anaerobic arginine degradation and nitrate respiration in Pseudomonas aeruginosa. J Bacteriol. 1991 Mar;173(5):1598–1606. doi: 10.1128/jb.173.5.1598-1606.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green J., Sharrocks A. D., Green B., Geisow M., Guest J. R. Properties of FNR proteins substituted at each of the five cysteine residues. Mol Microbiol. 1993 Apr;8(1):61–68. doi: 10.1111/j.1365-2958.1993.tb01203.x. [DOI] [PubMed] [Google Scholar]
- Gribskov M., Devereux J., Burgess R. R. The codon preference plot: graphic analysis of protein coding sequences and prediction of gene expression. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 2):539–549. doi: 10.1093/nar/12.1part2.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawley D. K., McClure W. R. Compilation and analysis of Escherichia coli promoter DNA sequences. Nucleic Acids Res. 1983 Apr 25;11(8):2237–2255. doi: 10.1093/nar/11.8.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoitink C. W., Woudt L. P., Turenhout J. C., van de Kamp M., Canters G. W. Isolation and sequencing of the Alcaligenes denitrificans azurin-encoding gene: comparison with the genes encoding blue copper proteins from Pseudomonas aeruginosa and Alcaligenes faecalis. Gene. 1990 May 31;90(1):15–20. doi: 10.1016/0378-1119(90)90434-s. [DOI] [PubMed] [Google Scholar]
- Iuchi S., Matsuda Z., Fujiwara T., Lin E. C. The arcB gene of Escherichia coli encodes a sensor-regulator protein for anaerobic repression of the arc modulon. Mol Microbiol. 1990 May;4(5):715–727. doi: 10.1111/j.1365-2958.1990.tb00642.x. [DOI] [PubMed] [Google Scholar]
- Jüngst A., Wakabayashi S., Matsubara H., Zumft W. G. The nirSTBM region coding for cytochrome cd1-dependent nitrite respiration of Pseudomonas stutzeri consists of a cluster of mono-, di-, and tetraheme proteins. FEBS Lett. 1991 Feb 25;279(2):205–209. doi: 10.1016/0014-5793(91)80150-2. [DOI] [PubMed] [Google Scholar]
- Jüngst A., Zumft W. G. Interdependence of respiratory NO reduction and nitrite reduction revealed by mutagenesis of nirQ, a novel gene in the denitrification gene cluster of Pseudomonas stutzeri. FEBS Lett. 1992 Dec 21;314(3):308–314. doi: 10.1016/0014-5793(92)81495-8. [DOI] [PubMed] [Google Scholar]
- Körner H., Frunzke K., Döhler K., Zumft W. G. Immunochemical patterns of distribution of nitrous oxide reductase and nitrite reductase (cytochrome cd1) among denitrifying pseudomonads. Arch Microbiol. 1987 Jun;148(1):20–24. doi: 10.1007/BF00429641. [DOI] [PubMed] [Google Scholar]
- Körner H., Zumft W. G. Expression of denitrification enzymes in response to the dissolved oxygen level and respiratory substrate in continuous culture of Pseudomonas stutzeri. Appl Environ Microbiol. 1989 Jul;55(7):1670–1676. doi: 10.1128/aem.55.7.1670-1676.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lodge J., Williams R., Bell A., Chan B., Busby S. Comparison of promoter activities in Escherichia coli and Pseudomonas aeruginosa: use of a new broad-host-range promoter-probe plasmid. FEMS Microbiol Lett. 1990 Jan 15;55(1-2):221–225. doi: 10.1016/0378-1097(90)90199-z. [DOI] [PubMed] [Google Scholar]
- Lüthi E., Mercenier A., Haas D. The arcABC operon required for fermentative growth of Pseudomonas aeruginosa on arginine: Tn5-751-assisted cloning and localization of structural genes. J Gen Microbiol. 1986 Oct;132(10):2667–2675. doi: 10.1099/00221287-132-10-2667. [DOI] [PubMed] [Google Scholar]
- MacGregor C. H., Wolff J. A., Arora S. K., Phibbs P. V., Jr Cloning of a catabolite repression control (crc) gene from Pseudomonas aeruginosa, expression of the gene in Escherichia coli, and identification of the gene product in Pseudomonas aeruginosa. J Bacteriol. 1991 Nov;173(22):7204–7212. doi: 10.1128/jb.173.22.7204-7212.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacInnes J. I., Kim J. E., Lian C. J., Soltes G. A. Actinobacillus pleuropneumoniae hlyX gene homology with the fnr gene of Escherichia coli. J Bacteriol. 1990 Aug;172(8):4587–4592. doi: 10.1128/jb.172.8.4587-4592.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maizel J. V., Jr, Lenk R. P. Enhanced graphic matrix analysis of nucleic acid and protein sequences. Proc Natl Acad Sci U S A. 1981 Dec;78(12):7665–7669. doi: 10.1073/pnas.78.12.7665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsubara T., Frunzke K., Zumft W. G. Modulation by copper of the products of nitrite respiration in Pseudomonas perfectomarinus. J Bacteriol. 1982 Mar;149(3):816–823. doi: 10.1128/jb.149.3.816-823.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordling M., Young S., Karlsson B. G., Lundberg L. G. The structural gene for cytochrome c551 from Pseudomonas aeruginosa. The nucleotide sequence shows a location downstream of the nitrite reductase gene. FEBS Lett. 1990 Jan 1;259(2):230–232. doi: 10.1016/0014-5793(90)80015-b. [DOI] [PubMed] [Google Scholar]
- Oka A., Sugisaki H., Takanami M. Nucleotide sequence of the kanamycin resistance transposon Tn903. J Mol Biol. 1981 Apr 5;147(2):217–226. doi: 10.1016/0022-2836(81)90438-1. [DOI] [PubMed] [Google Scholar]
- Römermann D., Warrelmann J., Bender R. A., Friedrich B. An rpoN-like gene of Alcaligenes eutrophus and Pseudomonas facilis controls expression of diverse metabolic pathways, including hydrogen oxidation. J Bacteriol. 1989 Feb;171(2):1093–1099. doi: 10.1128/jb.171.2.1093-1099.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savioz A., Zimmermann A., Haas D. Pseudomonas aeruginosa promoters which contain a conserved GG-N10-GC motif but appear to be RpoN-independent. Mol Gen Genet. 1993 Apr;238(1-2):74–80. doi: 10.1007/BF00279533. [DOI] [PubMed] [Google Scholar]
- Sawers G., Suppmann B. Anaerobic induction of pyruvate formate-lyase gene expression is mediated by the ArcA and FNR proteins. J Bacteriol. 1992 Jun;174(11):3474–3478. doi: 10.1128/jb.174.11.3474-3478.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawers R. G. Identification and molecular characterization of a transcriptional regulator from Pseudomonas aeruginosa PAO1 exhibiting structural and functional similarity to the FNR protein of Escherichia coli. Mol Microbiol. 1991 Jun;5(6):1469–1481. doi: 10.1111/j.1365-2958.1991.tb00793.x. [DOI] [PubMed] [Google Scholar]
- Sharrocks A. D., Green J., Guest J. R. FNR activates and represses transcription in vitro. Proc Biol Sci. 1991 Sep 23;245(1314):219–226. doi: 10.1098/rspb.1991.0113. [DOI] [PubMed] [Google Scholar]
- Simon R., O'Connell M., Labes M., Pühler A. Plasmid vectors for the genetic analysis and manipulation of rhizobia and other gram-negative bacteria. Methods Enzymol. 1986;118:640–659. doi: 10.1016/0076-6879(86)18106-7. [DOI] [PubMed] [Google Scholar]
- Smith G. B., Tiedje J. M. Isolation and characterization of a nitrite reductase gene and its use as a probe for denitrifying bacteria. Appl Environ Microbiol. 1992 Jan;58(1):376–384. doi: 10.1128/aem.58.1.376-384.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiro S. An FNR-dependent promoter from Escherichia coli is active and anaerobically inducible in Paracoccus denitrificans. FEMS Microbiol Lett. 1992 Nov 1;77(1-3):145–148. doi: 10.1016/0378-1097(92)90146-f. [DOI] [PubMed] [Google Scholar]
- Spiro S., Gaston K. L., Bell A. I., Roberts R. E., Busby S. J., Guest J. R. Interconversion of the DNA-binding specificities of two related transcription regulators, CRP and FNR. Mol Microbiol. 1990 Nov;4(11):1831–1838. doi: 10.1111/j.1365-2958.1990.tb02031.x. [DOI] [PubMed] [Google Scholar]
- Spiro S., Guest J. R. FNR and its role in oxygen-regulated gene expression in Escherichia coli. FEMS Microbiol Rev. 1990 Aug;6(4):399–428. doi: 10.1111/j.1574-6968.1990.tb04109.x. [DOI] [PubMed] [Google Scholar]
- Spiro S., Guest J. R. Regulation and over-expression of the fnr gene of Escherichia coli. J Gen Microbiol. 1987 Dec;133(12):3279–3288. doi: 10.1099/00221287-133-12-3279. [DOI] [PubMed] [Google Scholar]
- Tinoco I., Jr, Borer P. N., Dengler B., Levin M. D., Uhlenbeck O. C., Crothers D. M., Bralla J. Improved estimation of secondary structure in ribonucleic acids. Nat New Biol. 1973 Nov 14;246(150):40–41. doi: 10.1038/newbio246040a0. [DOI] [PubMed] [Google Scholar]
- Totten P. A., Lara J. C., Lory S. The rpoN gene product of Pseudomonas aeruginosa is required for expression of diverse genes, including the flagellin gene. J Bacteriol. 1990 Jan;172(1):389–396. doi: 10.1128/jb.172.1.389-396.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trageser M., Unden G. Role of cysteine residues and of metal ions in the regulatory functioning of FNR, the transcriptional regulator of anaerobic respiration in Escherichia coli. Mol Microbiol. 1989 May;3(5):593–599. doi: 10.1111/j.1365-2958.1989.tb00206.x. [DOI] [PubMed] [Google Scholar]
- Viebrock A., Zumft W. G. Molecular cloning, heterologous expression, and primary structure of the structural gene for the copper enzyme nitrous oxide reductase from denitrifying Pseudomonas stutzeri. J Bacteriol. 1988 Oct;170(10):4658–4668. doi: 10.1128/jb.170.10.4658-4668.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira J., Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982 Oct;19(3):259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- Weber I. T., Steitz T. A. Structure of a complex of catabolite gene activator protein and cyclic AMP refined at 2.5 A resolution. J Mol Biol. 1987 Nov 20;198(2):311–326. doi: 10.1016/0022-2836(87)90315-9. [DOI] [PubMed] [Google Scholar]
- Williams R., Bell A., Sims G., Busby S. The role of two surface exposed loops in transcription activation by the Escherichia coli CRP and FNR proteins. Nucleic Acids Res. 1991 Dec 25;19(24):6705–6712. doi: 10.1093/nar/19.24.6705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye R. W., Fries M. R., Bezborodnikov S. G., Averill B. A., Tiedje J. M. Characterization of the structural gene encoding a copper-containing nitrite reductase and homology of this gene to DNA of other denitrifiers. Appl Environ Microbiol. 1993 Jan;59(1):250–254. doi: 10.1128/aem.59.1.250-254.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann A., Reimmann C., Galimand M., Haas D. Anaerobic growth and cyanide synthesis of Pseudomonas aeruginosa depend on anr, a regulatory gene homologous with fnr of Escherichia coli. Mol Microbiol. 1991 Jun;5(6):1483–1490. doi: 10.1111/j.1365-2958.1991.tb00794.x. [DOI] [PubMed] [Google Scholar]
- Zumft W. G., Dreusch A., Löchelt S., Cuypers H., Friedrich B., Schneider B. Derived amino acid sequences of the nosZ gene (respiratory N2O reductase) from Alcaligenes eutrophus, Pseudomonas aeruginosa and Pseudomonas stutzeri reveal potential copper-binding residues. Implications for the CuA site of N2O reductase and cytochrome-c oxidase. Eur J Biochem. 1992 Aug 15;208(1):31–40. doi: 10.1111/j.1432-1033.1992.tb17156.x. [DOI] [PubMed] [Google Scholar]
- Zumft W. G., Döhler K., Körner H. Isolation and characterization of transposon Tn5-induced mutants of Pseudomonas perfectomarina defective in nitrous oxide respiration. J Bacteriol. 1985 Sep;163(3):918–924. doi: 10.1128/jb.163.3.918-924.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]