Abstract
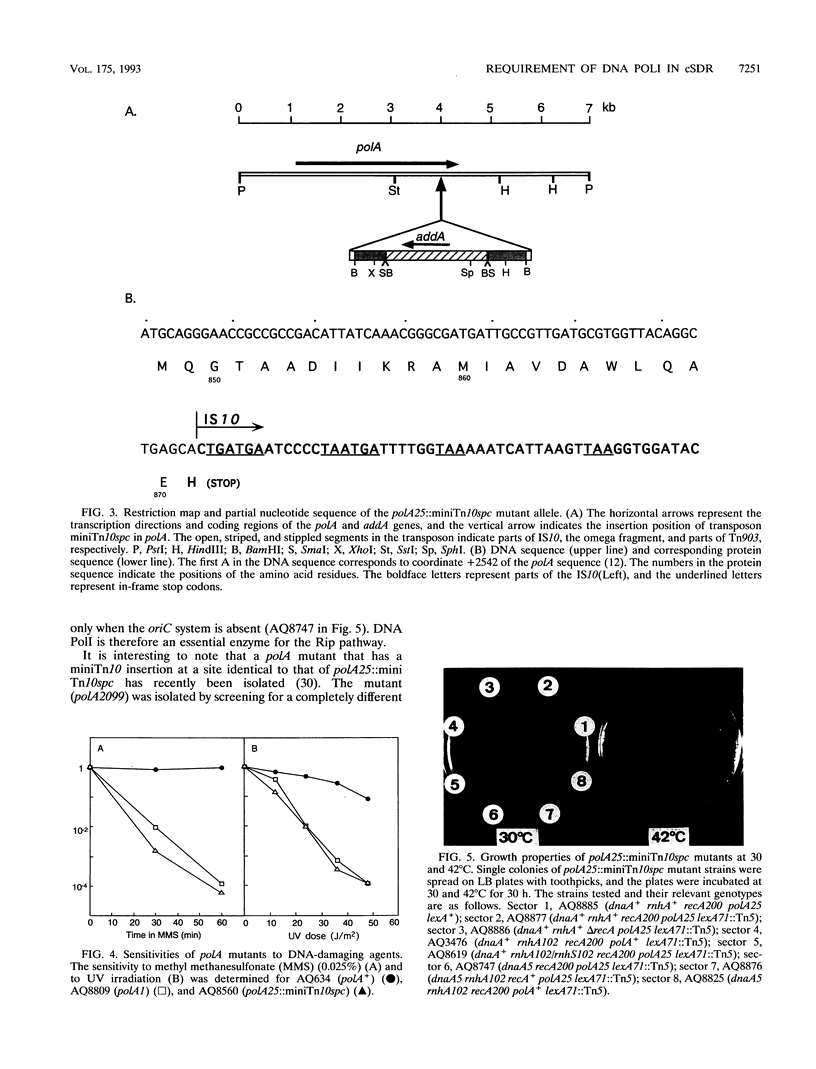
In Escherichia coli rnhA mutants, several normally repressed origins (oriK sites) of DNA replication are activated. The type of DNA replication initiated from these origins, termed constitutive stable DNA replication, does not require DnaA protein or the oriC site, which are essential for normal DNA replication. It requires active RecA protein. We previously found that the lexA71(Def)::Tn5 mutation can suppress this RecA requirement and postulated that the derepression of a LexA regulon gene(s) leads to the activation of a bypass pathway, Rip (for RecA-independent process). In this study, we isolated a miniTn10spc insertion mutant that abolishes the ability of the lexA(Def) mutation to suppress the RecA requirement of constitutive stable DNA replication. Cloning and DNA sequencing analysis of the mutant revealed that the insertion occurs at the 3' end of the coding region of the polA gene, which encodes DNA polymerase I. The mutant allele, designated polA25::miniTn10spc, is expected to abolish the polymerization activity but not the 5'-->3' or 3'-->5' exonuclease activity. Thus, the Rip bypass pathway requires active DNA polymerase I. Since the lethal combination of recA(Def) and polA25::miniTn10spc could be suppressed by derepression of the LexA regulon only when DNA replication is driven by the oriC system, it was suggested that the bypass pathway has a specific requirement for DNA polymerase I at the initiation step in the absence of RecA. An accompanying paper (Y. Cao and T. Kogoma, J. Bacteriol. 175:7254-7259, 1993) describes experiments to determine which activities of DNA polymerase I are required at the initiation step and discusses possible roles for DNA polymerase in the Rip bypass pathway.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Biek D. P., Cohen S. N. Identification and characterization of recD, a gene affecting plasmid maintenance and recombination in Escherichia coli. J Bacteriol. 1986 Aug;167(2):594–603. doi: 10.1128/jb.167.2.594-603.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochner B. R., Huang H. C., Schieven G. L., Ames B. N. Positive selection for loss of tetracycline resistance. J Bacteriol. 1980 Aug;143(2):926–933. doi: 10.1128/jb.143.2.926-933.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonner C. A., Hays S., McEntee K., Goodman M. F. DNA polymerase II is encoded by the DNA damage-inducible dinA gene of Escherichia coli. Proc Natl Acad Sci U S A. 1990 Oct;87(19):7663–7667. doi: 10.1073/pnas.87.19.7663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y., Kogoma T. Requirement for the polymerization and 5'-->3' exonuclease activities of DNA polymerase I in initiation of DNA replication at oriK sites in the absence of RecA in Escherichia coli rnhA mutants. J Bacteriol. 1993 Nov;175(22):7254–7259. doi: 10.1128/jb.175.22.7254-7259.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadaban M. J., Cohen S. N. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J Mol Biol. 1980 Apr;138(2):179–207. doi: 10.1016/0022-2836(80)90283-1. [DOI] [PubMed] [Google Scholar]
- Fijalkowska I., Jonczyk P., Ciesla Z. Conditional lethality of the recA441 and recA730 mutants of Escherichia coli deficient in DNA polymerase I. Mutat Res. 1989 Mar;217(2):117–122. doi: 10.1016/0921-8777(89)90063-3. [DOI] [PubMed] [Google Scholar]
- Gross J. D., Grunstein J., Witkin E. M. Inviability of recA- derivatives of the DNA polymerase mutant of De Lucia and Cairns. J Mol Biol. 1971 Jun 14;58(2):631–634. doi: 10.1016/0022-2836(71)90377-9. [DOI] [PubMed] [Google Scholar]
- Halling S. M., Kleckner N. A symmetrical six-base-pair target site sequence determines Tn10 insertion specificity. Cell. 1982 Jan;28(1):155–163. doi: 10.1016/0092-8674(82)90385-3. [DOI] [PubMed] [Google Scholar]
- Irino N., Nakayama K., Nakayama H. The recQ gene of Escherichia coli K12: primary structure and evidence for SOS regulation. Mol Gen Genet. 1986 Nov;205(2):298–304. doi: 10.1007/BF00430442. [DOI] [PubMed] [Google Scholar]
- Iwasaki H., Nakata A., Walker G. C., Shinagawa H. The Escherichia coli polB gene, which encodes DNA polymerase II, is regulated by the SOS system. J Bacteriol. 1990 Nov;172(11):6268–6273. doi: 10.1128/jb.172.11.6268-6273.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyce C. M., Kelley W. S., Grindley N. D. Nucleotide sequence of the Escherichia coli polA gene and primary structure of DNA polymerase I. J Biol Chem. 1982 Feb 25;257(4):1958–1964. [PubMed] [Google Scholar]
- Kaasch M., Kaasch J., Quiñones A. Expression of the dnaN and dnaQ genes of Escherichia coli is inducible by mitomycin C. Mol Gen Genet. 1989 Oct;219(1-2):187–192. doi: 10.1007/BF00261175. [DOI] [PubMed] [Google Scholar]
- Karran P., Lindahl T., Ofsteng I., Evensen G. B., Seeberg E. Escherichia coli mutants deficient in 3-methyladenine-DNA glycosylase. J Mol Biol. 1980 Jun 15;140(1):101–127. doi: 10.1016/0022-2836(80)90358-7. [DOI] [PubMed] [Google Scholar]
- Kelley W. S. Mapping of the polA locus of Escherichia coli K12: genetic fine structure of the cistron. Genetics. 1980 May;95(1):15–38. doi: 10.1093/genetics/95.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kogoma T. Absence of RNase H allows replication of pBR322 in Escherichia coli mutants lacking DNA polymerase I. Proc Natl Acad Sci U S A. 1984 Dec;81(24):7845–7849. doi: 10.1073/pnas.81.24.7845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kogoma T., Skarstad K., Boye E., von Meyenburg K., Steen H. B. RecA protein acts at the initiation of stable DNA replication in rnh mutants of Escherichia coli K-12. J Bacteriol. 1985 Aug;163(2):439–444. doi: 10.1128/jb.163.2.439-444.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kogoma T., von Meyenburg K. The origin of replication, oriC, and the dnaA protein are dispensable in stable DNA replication (sdrA) mutants of Escherichia coli K-12. EMBO J. 1983;2(3):463–468. doi: 10.1002/j.1460-2075.1983.tb01445.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohara Y., Akiyama K., Isono K. The physical map of the whole E. coli chromosome: application of a new strategy for rapid analysis and sorting of a large genomic library. Cell. 1987 Jul 31;50(3):495–508. doi: 10.1016/0092-8674(87)90503-4. [DOI] [PubMed] [Google Scholar]
- LENNOX E. S. Transduction of linked genetic characters of the host by bacteriophage P1. Virology. 1955 Jul;1(2):190–206. doi: 10.1016/0042-6822(55)90016-7. [DOI] [PubMed] [Google Scholar]
- Lloyd R. G., Barbour S. D. The genetic location of the sbcA gene of Escherichia coli. Mol Gen Genet. 1974;134(2):157–171. doi: 10.1007/BF00268417. [DOI] [PubMed] [Google Scholar]
- Lloyd R. G., Buckman C. Genetic analysis of the recG locus of Escherichia coli K-12 and of its role in recombination and DNA repair. J Bacteriol. 1991 Feb;173(3):1004–1011. doi: 10.1128/jb.173.3.1004-1011.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd R. G., Picksley S. M., Prescott C. Inducible expression of a gene specific to the RecF pathway for recombination in Escherichia coli K12. Mol Gen Genet. 1983;190(1):162–167. doi: 10.1007/BF00330340. [DOI] [PubMed] [Google Scholar]
- Monk M., Kinross J. Conditional lethality of recA and recB derivatives of a strain of Escherichia coli K-12 with a temperature-sensitive deoxyribonucleic acid polymerase I. J Bacteriol. 1972 Mar;109(3):971–978. doi: 10.1128/jb.109.3.971-978.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimyo M., Shimazu Y. Evidence that the gene uvrB is indispensable for a polymerase I deficient strain of Escherichia coli K-12. Mol Gen Genet. 1976 Sep 23;147(3):243–250. doi: 10.1007/BF00582875. [DOI] [PubMed] [Google Scholar]
- Ogawa T., Pickett G. G., Kogoma T., Kornberg A. RNase H confers specificity in the dnaA-dependent initiation of replication at the unique origin of the Escherichia coli chromosome in vivo and in vitro. Proc Natl Acad Sci U S A. 1984 Feb;81(4):1040–1044. doi: 10.1073/pnas.81.4.1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prentki P., Binda A., Epstein A. Plasmid vectors for selecting IS1-promoted deletions in cloned DNA: sequence analysis of the omega interposon. Gene. 1991 Jul 15;103(1):17–23. doi: 10.1016/0378-1119(91)90385-o. [DOI] [PubMed] [Google Scholar]
- Quiñones A., Kaasch J., Kaasch M., Messer W. Induction of dnaN and dnaQ gene expression in Escherichia coli by alkylation damage to DNA. EMBO J. 1989 Feb;8(2):587–593. doi: 10.1002/j.1460-2075.1989.tb03413.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro J. A. Differential action and differential expression of DNA polymerase I during Escherichia coli colony development. J Bacteriol. 1992 Nov;174(22):7262–7272. doi: 10.1128/jb.174.22.7262-7272.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shurvinton C. E., Lloyd R. G. Damage to DNA induces expression of the ruv gene of Escherichia coli. Mol Gen Genet. 1982;185(2):352–355. doi: 10.1007/BF00330811. [DOI] [PubMed] [Google Scholar]
- Singer M., Baker T. A., Schnitzler G., Deischel S. M., Goel M., Dove W., Jaacks K. J., Grossman A. D., Erickson J. W., Gross C. A. A collection of strains containing genetically linked alternating antibiotic resistance elements for genetic mapping of Escherichia coli. Microbiol Rev. 1989 Mar;53(1):1–24. doi: 10.1128/mr.53.1.1-24.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subia N. L., Kogoma T. Concatemer formation of ColE1-type plasmids in mutants of Escherichia coli lacking RNase H activity. J Mol Biol. 1986 Jun 5;189(3):389–399. doi: 10.1016/0022-2836(86)90311-6. [DOI] [PubMed] [Google Scholar]
- Sweasy J. B., Loeb L. A. Mammalian DNA polymerase beta can substitute for DNA polymerase I during DNA replication in Escherichia coli. J Biol Chem. 1992 Jan 25;267(3):1407–1410. [PubMed] [Google Scholar]
- Torrey T. A., Kogoma T. Genetic analysis of constitutive stable DNA replication in rnh mutants of Escherichia coli K12. Mol Gen Genet. 1987 Jul;208(3):420–427. doi: 10.1007/BF00328133. [DOI] [PubMed] [Google Scholar]
- Torrey T. A., Kogoma T. Suppressor mutations (rin) that specifically suppress the recA+ dependence of stable DNA replication in Escherichia coliK-12. Mol Gen Genet. 1982;187(2):225–230. doi: 10.1007/BF00331121. [DOI] [PubMed] [Google Scholar]
- Walker G. C. Mutagenesis and inducible responses to deoxyribonucleic acid damage in Escherichia coli. Microbiol Rev. 1984 Mar;48(1):60–93. doi: 10.1128/mr.48.1.60-93.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward D. F., Murray N. E. Construction and characterization of Escherichia coli polA-lacZ gene fusions. J Bacteriol. 1980 Jun;142(3):962–972. doi: 10.1128/jb.142.3.962-972.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Way J. C., Davis M. A., Morisato D., Roberts D. E., Kleckner N. New Tn10 derivatives for transposon mutagenesis and for construction of lacZ operon fusions by transposition. Gene. 1984 Dec;32(3):369–379. doi: 10.1016/0378-1119(84)90012-x. [DOI] [PubMed] [Google Scholar]
- Wilson G. G., Young K. Y., Edlin G. J., Konigsberg W. High-frequency generalised transduction by bacteriophage T4. Nature. 1979 Jul 5;280(5717):80–82. doi: 10.1038/280080a0. [DOI] [PubMed] [Google Scholar]
- Witkin E. M. RecA protein in the SOS response: milestones and mysteries. Biochimie. 1991 Feb-Mar;73(2-3):133–141. doi: 10.1016/0300-9084(91)90196-8. [DOI] [PubMed] [Google Scholar]
- Witkin E. M., Roegner-Maniscalco V. Overproduction of DnaE protein (alpha subunit of DNA polymerase III) restores viability in a conditionally inviable Escherichia coli strain deficient in DNA polymerase I. J Bacteriol. 1992 Jun;174(12):4166–4168. doi: 10.1128/jb.174.12.4166-4168.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- de Massy B., Fayet O., Kogoma T. Multiple origin usage for DNA replication in sdrA(rnh) mutants of Escherichia coli K-12. Initiation in the absence of oriC. J Mol Biol. 1984 Sep 15;178(2):227–236. doi: 10.1016/0022-2836(84)90141-4. [DOI] [PubMed] [Google Scholar]
- von Meyenburg K., Boye E., Skarstad K., Koppes L., Kogoma T. Mode of initiation of constitutive stable DNA replication in RNase H-defective mutants of Escherichia coli K-12. J Bacteriol. 1987 Jun;169(6):2650–2658. doi: 10.1128/jb.169.6.2650-2658.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]