Abstract
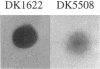
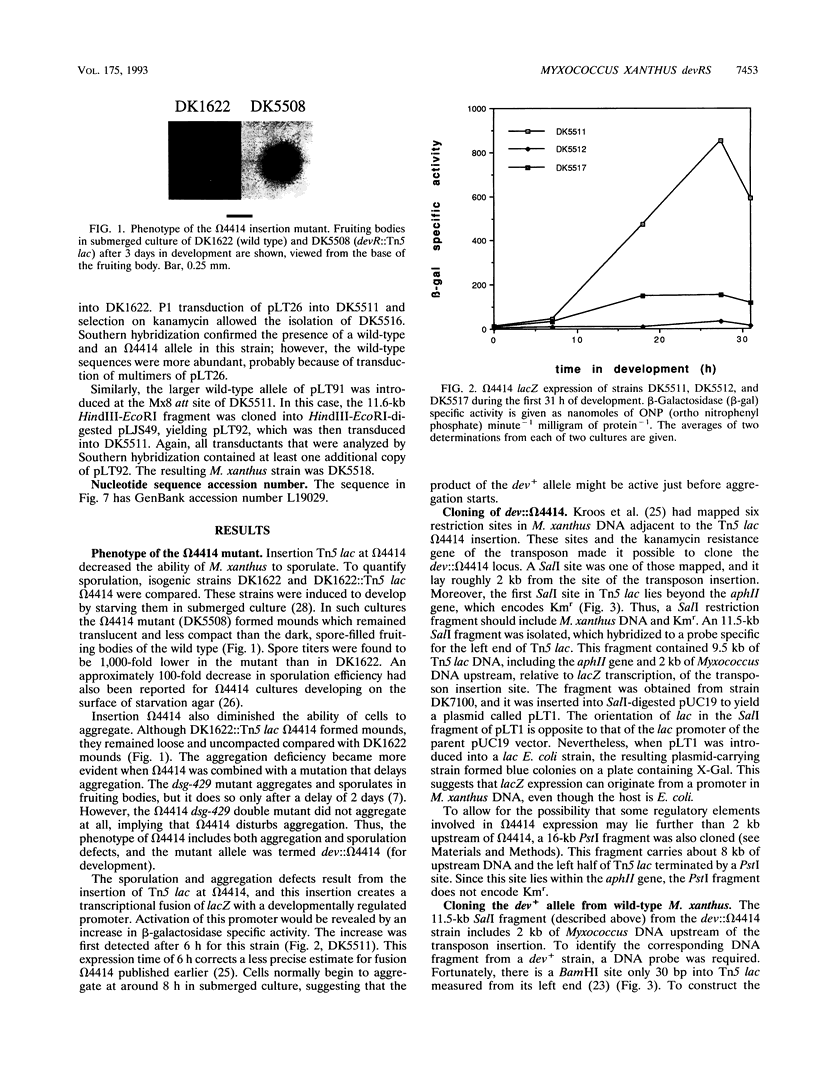
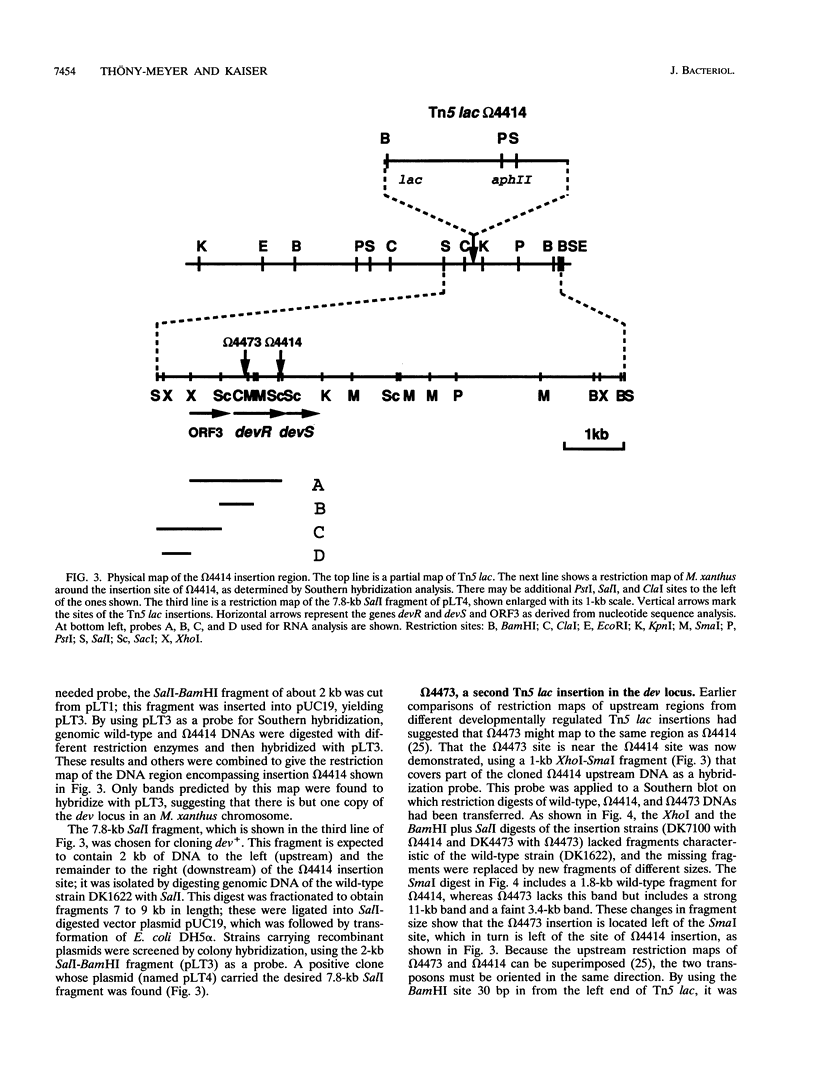
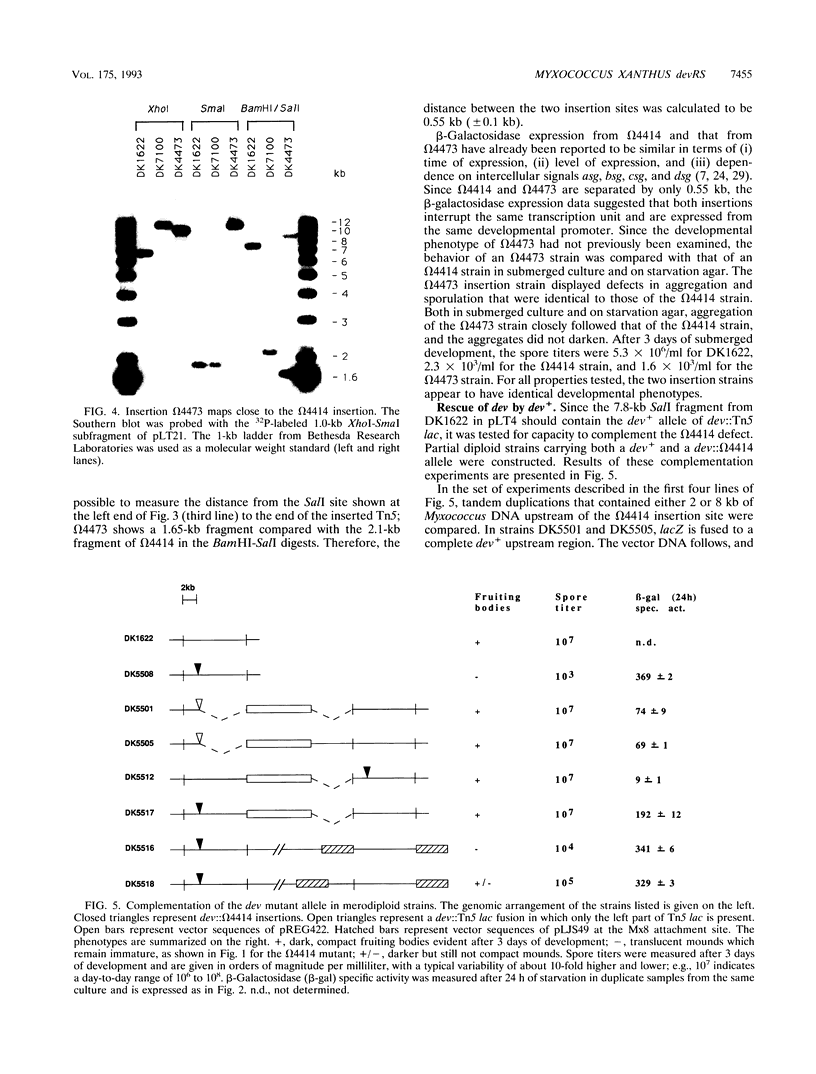
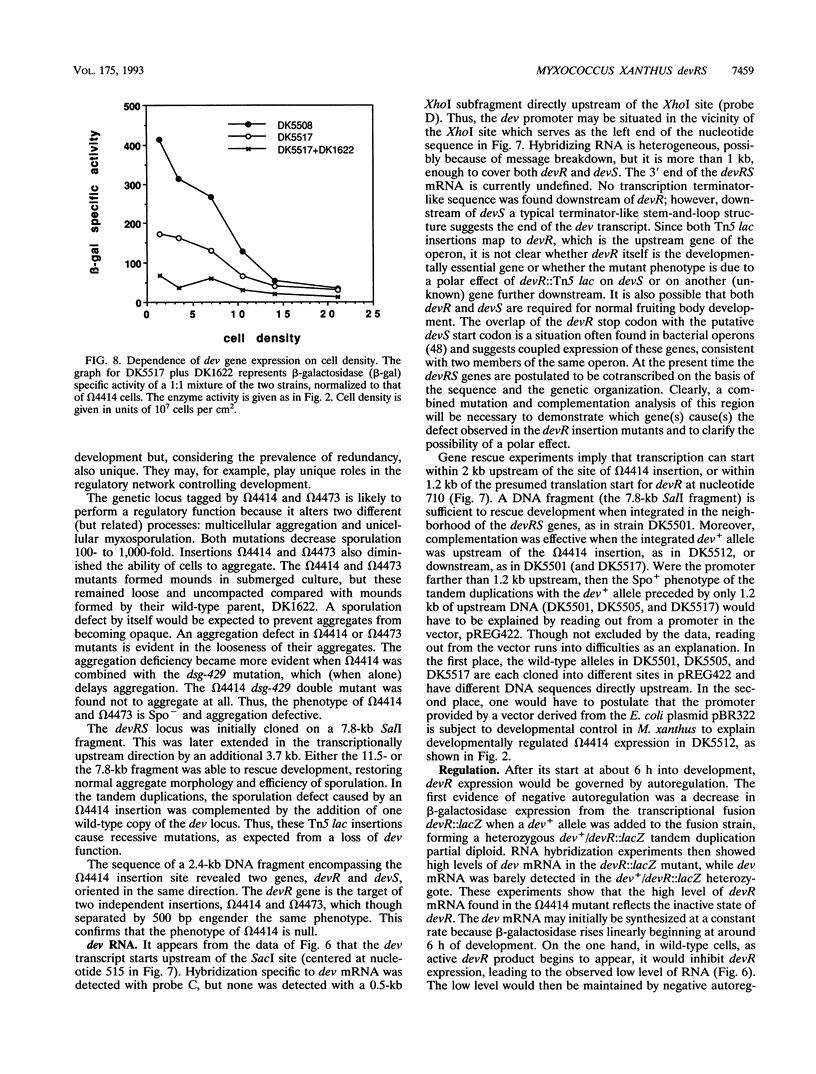
Two Tn5 lac insertions into the Myxococcus genome at sites omega 4414 and omega 4473, which are separated by 550 nucleotides, inactivate fruiting body development. Sporulation is decreased 100- to 10,000-fold. At least two genes, devR and devS, are transcribed in this region, probably as an operon. Expression of devR begins by 6 h after starvation has initiated development. On the basis of their nucleotide sequences, devR and devS are expected to encode proteins of 302 and 214 amino acids, respectively. Dev+ function can be restored by a segment of 7.8 kb cloned from the devRS region of wild-type cells. Two experiments show that devR expression is under strong negative autoregulation. beta-Galactosidase is expressed at a higher level from a transcriptional devR::lacZ fusion when the fused operon is in a dev strain than when it is in the dev/dev+ genetic background of a partial diploid. There is more mRNA accumulation from the devRS region in the dev strain than in a rescued dev/dev+ tandem duplication strain. Sporulation rescue is correlated with some degree of negative autoregulation, even though sporulation is not inversely proportional to beta-galactosidase expression from omega 4414. A second level of regulation is suggested by complementation of dev by dev+ in duplication strains. The expression of devRS, measured by sporulation levels, differs 1,000-fold when devRS+ is moved from a distance of 20 kb to 3 Mb from the mutant devRS locus. Expression of devR is also dependent on the cell density at which development is initiated, a third level of regulation. Multiple levels of regulation suggest that devRS is a switch required to activate completion of aggregation and sporulation.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Apelian D., Inouye S. Development-specific sigma-factor essential for late-stage differentiation of Myxococcus xanthus. Genes Dev. 1990 Aug;4(8):1396–1403. doi: 10.1101/gad.4.8.1396. [DOI] [PubMed] [Google Scholar]
- Avery L., Kaiser D. In situ transposon replacement and isolation of a spontaneous tandem genetic duplication. Mol Gen Genet. 1983;191(1):99–109. doi: 10.1007/BF00330896. [DOI] [PubMed] [Google Scholar]
- Bibb M. J., Bibb M. J., Ward J. M., Cohen S. N. Nucleotide sequences encoding and promoting expression of three antibiotic resistance genes indigenous to Streptomyces. Mol Gen Genet. 1985;199(1):26–36. doi: 10.1007/BF00327505. [DOI] [PubMed] [Google Scholar]
- Bolivar F., Rodriguez R. L., Greene P. J., Betlach M. C., Heyneker H. L., Boyer H. W., Crosa J. H., Falkow S. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2(2):95–113. [PubMed] [Google Scholar]
- Casadaban M. J., Martinez-Arias A., Shapira S. K., Chou J. Beta-galactosidase gene fusions for analyzing gene expression in escherichia coli and yeast. Methods Enzymol. 1983;100:293–308. doi: 10.1016/0076-6879(83)00063-4. [DOI] [PubMed] [Google Scholar]
- Chen H. W., Kuspa A., Keseler I. M., Shimkets L. J. Physical map of the Myxococcus xanthus chromosome. J Bacteriol. 1991 Mar;173(6):2109–2115. doi: 10.1128/jb.173.6.2109-2115.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y., Kaiser D. dsg, a gene required for cell-cell interaction early in Myxococcus development. J Bacteriol. 1989 Jul;171(7):3719–3726. doi: 10.1128/jb.171.7.3719-3726.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisselsoder J., Campos J. M., Zusman D. R. Physical characterization of bacteriophage MX4, a generalized transducing phage for Myxococcus xanthus. J Mol Biol. 1978 Feb 25;119(2):179–189. doi: 10.1016/0022-2836(78)90432-1. [DOI] [PubMed] [Google Scholar]
- Gill J. S., Dworkin M. Cell surface antigens during submerged development of Myxococcus xanthus examined with monoclonal antibodies. J Bacteriol. 1986 Nov;168(2):505–511. doi: 10.1128/jb.168.2.505-511.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill J. S., Jarvis B. W., Dworkin M. Inhibition of development in Myxococcus xanthus by monoclonal antibody 1604. Proc Natl Acad Sci U S A. 1987 Jul;84(13):4505–4508. doi: 10.1073/pnas.84.13.4505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill R. E., Cull M. G., Fly S. Genetic identification and cloning of a gene required for developmental cell interactions in Myxococcus xanthus. J Bacteriol. 1988 Nov;170(11):5279–5288. doi: 10.1128/jb.170.11.5279-5288.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenblatt J., Schleif R. Arabinose C protein: regulation of the arabinose operon in vitro. Nat New Biol. 1971 Oct 6;233(40):166–170. doi: 10.1038/newbio233166a0. [DOI] [PubMed] [Google Scholar]
- Inouye M., Inouye S., Zusman D. R. Biosynthesis and self-assembly of protein S, a development-specific protein of Myxococcus xanthus. Proc Natl Acad Sci U S A. 1979 Jan;76(1):209–213. doi: 10.1073/pnas.76.1.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inouye M., Inouye S., Zusman D. R. Gene expression during development of Myxococcus xanthus: pattern of protein synthesis. Dev Biol. 1979 Feb;68(2):579–591. doi: 10.1016/0012-1606(79)90228-8. [DOI] [PubMed] [Google Scholar]
- Inouye S., Franceschini T., Inouye M. Structural similarities between the development-specific protein S from a gram-negative bacterium, Myxococcus xanthus, and calmodulin. Proc Natl Acad Sci U S A. 1983 Nov;80(22):6829–6833. doi: 10.1073/pnas.80.22.6829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis B. W., Dworkin M. Role of Myxococcus xanthus cell surface antigen 1604 in development. J Bacteriol. 1989 Sep;171(9):4667–4673. doi: 10.1128/jb.171.9.4667-4673.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser D., Kroos L., Kuspa A. Cell interactions govern the temporal pattern of Myxococcus development. Cold Spring Harb Symp Quant Biol. 1985;50:823–830. doi: 10.1101/sqb.1985.050.01.100. [DOI] [PubMed] [Google Scholar]
- Kaplan H. B., Kuspa A., Kaiser D. Suppressors that permit A-signal-independent developmental gene expression in Myxococcus xanthus. J Bacteriol. 1991 Feb;173(4):1460–1470. doi: 10.1128/jb.173.4.1460-1470.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kil K. S., Brown G. L., Downard J. S. A segment of Myxococcus xanthus ops DNA functions as an upstream activation site for tps gene transcription. J Bacteriol. 1990 Jun;172(6):3081–3088. doi: 10.1128/jb.172.6.3081-3088.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S. K., Kaiser D. C-factor has distinct aggregation and sporulation thresholds during Myxococcus development. J Bacteriol. 1991 Mar;173(5):1722–1728. doi: 10.1128/jb.173.5.1722-1728.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S. K., Kaiser D. C-factor: a cell-cell signaling protein required for fruiting body morphogenesis of M. xanthus. Cell. 1990 Apr 6;61(1):19–26. doi: 10.1016/0092-8674(90)90211-v. [DOI] [PubMed] [Google Scholar]
- Komano T., Furuichi T., Teintze M., Inouye M., Inouye S. Effects of deletion of the gene for the development-specific protein S on differentiation in Myxococcus xanthus. J Bacteriol. 1984 Jun;158(3):1195–1197. doi: 10.1128/jb.158.3.1195-1197.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroos L., Kaiser D. Construction of Tn5 lac, a transposon that fuses lacZ expression to exogenous promoters, and its introduction into Myxococcus xanthus. Proc Natl Acad Sci U S A. 1984 Sep;81(18):5816–5820. doi: 10.1073/pnas.81.18.5816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroos L., Kaiser D. Expression of many developmentally regulated genes in Myxococcus depends on a sequence of cell interactions. Genes Dev. 1987 Oct;1(8):840–854. doi: 10.1101/gad.1.8.840. [DOI] [PubMed] [Google Scholar]
- Kroos L., Kuspa A., Kaiser D. A global analysis of developmentally regulated genes in Myxococcus xanthus. Dev Biol. 1986 Sep;117(1):252–266. doi: 10.1016/0012-1606(86)90368-4. [DOI] [PubMed] [Google Scholar]
- Kroos L., Kuspa A., Kaiser D. Defects in fruiting body development caused by Tn5 lac insertions in Myxococcus xanthus. J Bacteriol. 1990 Jan;172(1):484–487. doi: 10.1128/jb.172.1.484-487.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuner J. M., Kaiser D. Fruiting body morphogenesis in submerged cultures of Myxococcus xanthus. J Bacteriol. 1982 Jul;151(1):458–461. doi: 10.1128/jb.151.1.458-461.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuspa A., Kroos L., Kaiser D. Intercellular signaling is required for developmental gene expression in Myxococcus xanthus. Dev Biol. 1986 Sep;117(1):267–276. doi: 10.1016/0012-1606(86)90369-6. [DOI] [PubMed] [Google Scholar]
- Kuspa A., Loomis W. F. Tagging developmental genes in Dictyostelium by restriction enzyme-mediated integration of plasmid DNA. Proc Natl Acad Sci U S A. 1992 Sep 15;89(18):8803–8807. doi: 10.1073/pnas.89.18.8803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuspa A., Plamann L., Kaiser D. Identification of heat-stable A-factor from Myxococcus xanthus. J Bacteriol. 1992 May;174(10):3319–3326. doi: 10.1128/jb.174.10.3319-3326.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Lee B. U., Shimkets L. J. csgA expression entrains Myxococcus xanthus development. Genes Dev. 1992 Mar;6(3):401–410. doi: 10.1101/gad.6.3.401. [DOI] [PubMed] [Google Scholar]
- Maloy S., Stewart V. Autogenous regulation of gene expression. J Bacteriol. 1993 Jan;175(2):307–316. doi: 10.1128/jb.175.2.307-316.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin S., Sodergren E., Masuda T., Kaiser D. Systematic isolation of transducing phages for Myxococcus xanthus. Virology. 1978 Jul 1;88(1):44–53. doi: 10.1016/0042-6822(78)90108-3. [DOI] [PubMed] [Google Scholar]
- Muñoz-Dorado J., Inouye S., Inouye M. A gene encoding a protein serine/threonine kinase is required for normal development of M. xanthus, a gram-negative bacterium. Cell. 1991 Nov 29;67(5):995–1006. doi: 10.1016/0092-8674(91)90372-6. [DOI] [PubMed] [Google Scholar]
- Norrander J., Kempe T., Messing J. Construction of improved M13 vectors using oligodeoxynucleotide-directed mutagenesis. Gene. 1983 Dec;26(1):101–106. doi: 10.1016/0378-1119(83)90040-9. [DOI] [PubMed] [Google Scholar]
- Plamann L., Kuspa A., Kaiser D. Proteins that rescue A-signal-defective mutants of Myxococcus xanthus. J Bacteriol. 1992 May;174(10):3311–3318. doi: 10.1128/jb.174.10.3311-3318.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romeo J. M., Zusman D. R. Cloning of the gene for myxobacterial hemagglutinin and isolation and analysis of structural gene mutations. J Bacteriol. 1987 Aug;169(8):3801–3808. doi: 10.1128/jb.169.8.3801-3808.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo-Marie F., Roederer M., Sager B., Herzenberg L. A., Kaiser D. Beta-galactosidase activity in single differentiating bacterial cells. Proc Natl Acad Sci U S A. 1993 Sep 1;90(17):8194–8198. doi: 10.1073/pnas.90.17.8194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimkets L. J., Asher S. J. Use of recombination techniques to examine the structure of the csg locus of Myxococcus xanthus. Mol Gen Genet. 1988 Jan;211(1):63–71. doi: 10.1007/BF00338394. [DOI] [PubMed] [Google Scholar]
- Shimkets L. J. Social and developmental biology of the myxobacteria. Microbiol Rev. 1990 Dec;54(4):473–501. doi: 10.1128/mr.54.4.473-501.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stellwag E., Fink J. M., Zissler J. Physical characterization of the genome of the Myxococcus xanthus bacteriophage MX-8. Mol Gen Genet. 1985;199(1):123–132. doi: 10.1007/BF00327521. [DOI] [PubMed] [Google Scholar]
- Sun D. X., Cabrera-Martinez R. M., Setlow P. Control of transcription of the Bacillus subtilis spoIIIG gene, which codes for the forespore-specific transcription factor sigma G. J Bacteriol. 1991 May;173(9):2977–2984. doi: 10.1128/jb.173.9.2977-2984.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]