Abstract
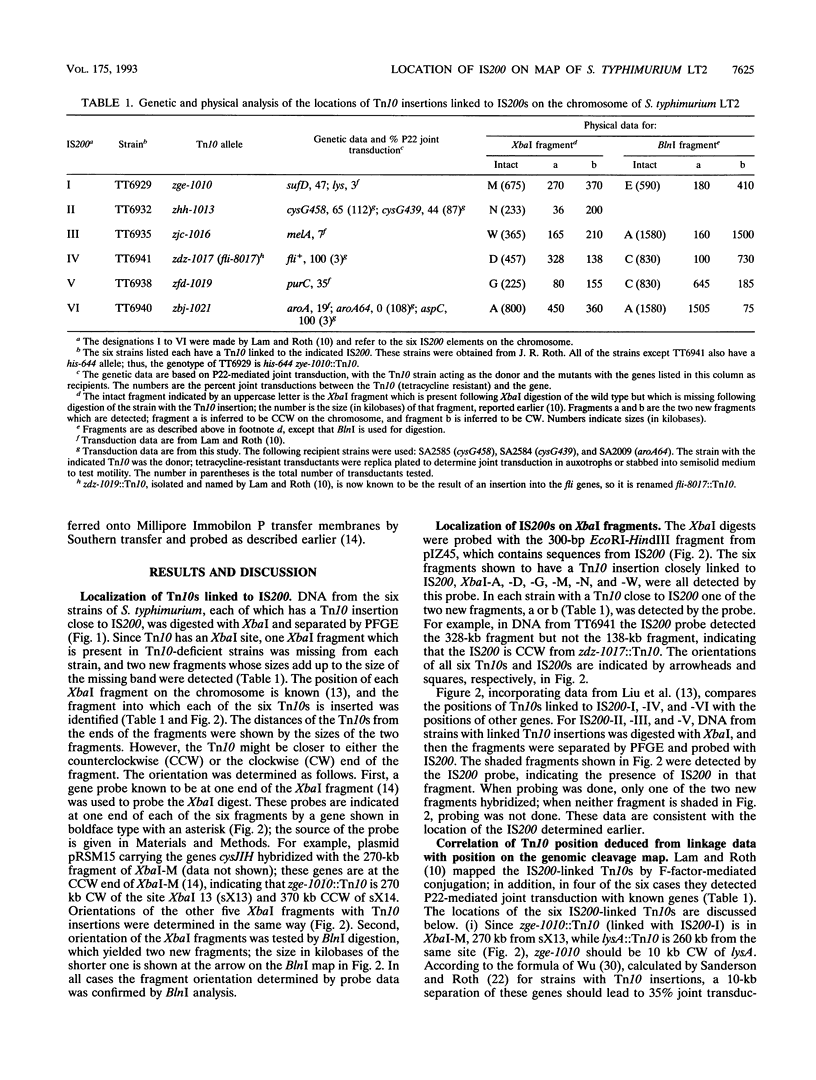
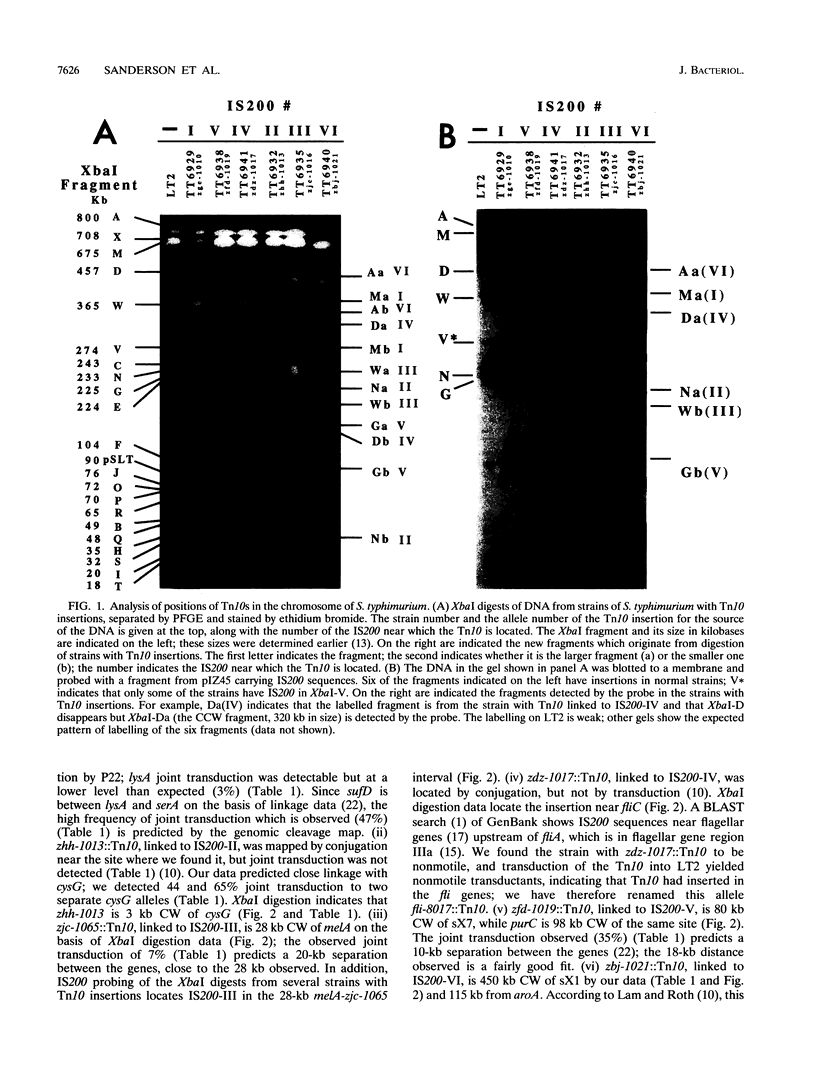
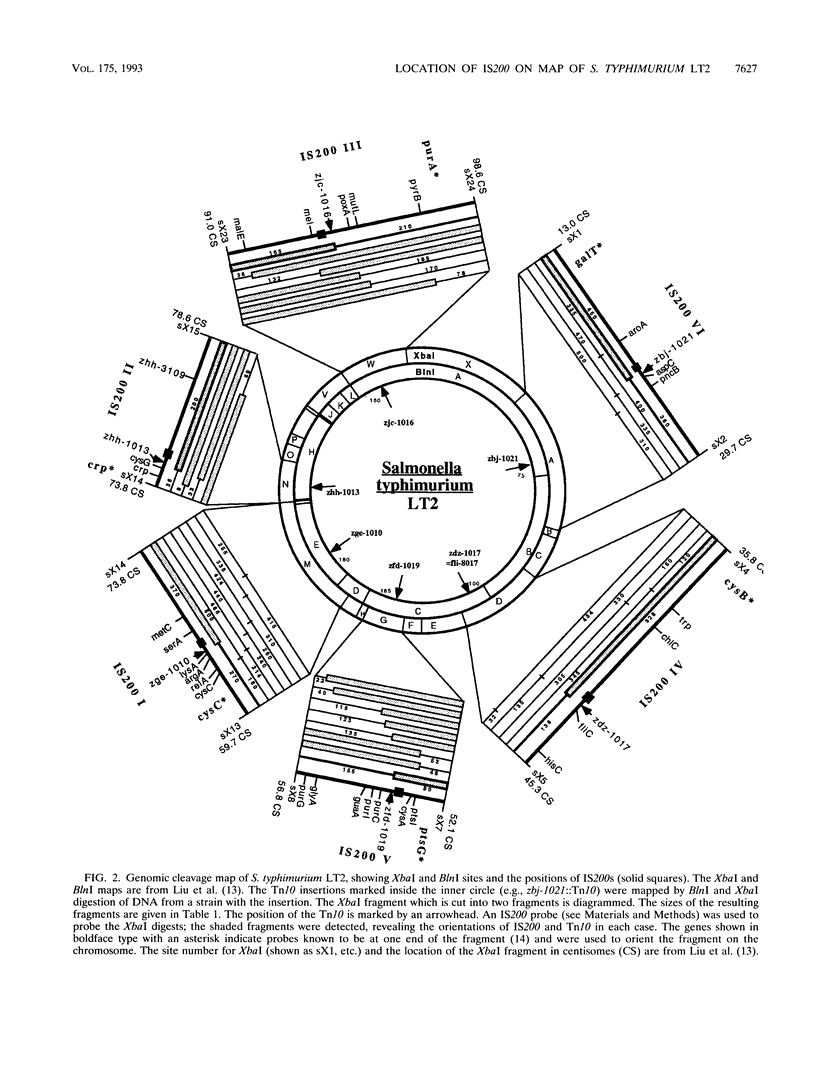
Locations of six Tn10s, closely linked to each of the six IS200s on the genomic cleavage map of Salmonella typhimurium LT2, were determined by digestion with XbaI and BlnI and separation of the fragments by pulsed-field gel electrophoresis; the locations were then further defined by P22-mediated joint transduction. The orientation of each IS200 with respect to its linked Tn10 was determined by Southern blotting. The locations of IS200-I, IS200-III, and IS200-V were confirmed to be close to sufD, melB, and purC, as previously indicated. IS200-II is jointly transduced with cysG. IS200-IV is near fliA; the linked Tn10 is inserted in fli, making the strain nonmotile. IS200-VI is jointly transduced with aspC but not with aroA. IS200 is transposed to a seventh site in some strains, while remaining in the other six locations described above. These data indicate that genome analysis by pulsed-field gel electrophoresis can locate the positions of Tn10s with accuracy sufficient to predict P22-mediated joint transduction.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. Basic local alignment search tool. J Mol Biol. 1990 Oct 5;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Bisercić M., Ochman H. Natural populations of Escherichia coli and Salmonella typhimurium harbor the same classes of insertion sequences. Genetics. 1993 Mar;133(3):449–454. doi: 10.1093/genetics/133.3.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadesus J., Roth J. R. Absence of insertions among spontaneous mutants of Salmonella typhimurium. Mol Gen Genet. 1989 Apr;216(2-3):210–216. doi: 10.1007/BF00334358. [DOI] [PubMed] [Google Scholar]
- Gibert I., Barbé J., Casadesús J. Distribution of insertion sequence IS200 in Salmonella and Shigella. J Gen Microbiol. 1990 Dec;136(12):2555–2560. doi: 10.1099/00221287-136-12-2555. [DOI] [PubMed] [Google Scholar]
- Gibert I., Carroll K., Hillyard D. R., Barbé J., Casadesus J. IS200 is not a member of the IS600 family of insertion sequences. Nucleic Acids Res. 1991 Mar 25;19(6):1343–1343. doi: 10.1093/nar/19.6.1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan E., Saedler H., Starlinger P. O0 and strong-polar mutations in the gal operon are insertions. Mol Gen Genet. 1968;102(4):353–363. doi: 10.1007/BF00433726. [DOI] [PubMed] [Google Scholar]
- Krawiec S., Riley M. Organization of the bacterial chromosome. Microbiol Rev. 1990 Dec;54(4):502–539. doi: 10.1128/mr.54.4.502-539.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam S., Roth J. R. Genetic mapping of IS200 copies in Salmonella typhimurim strain LT2. Genetics. 1983 Dec;105(4):801–811. doi: 10.1093/genetics/105.4.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam S., Roth J. R. IS200: a Salmonella-specific insertion sequence. Cell. 1983 Oct;34(3):951–960. doi: 10.1016/0092-8674(83)90552-4. [DOI] [PubMed] [Google Scholar]
- Lam S., Roth J. R. Structural and functional studies of insertion element IS200. J Mol Biol. 1986 Jan 20;187(2):157–167. doi: 10.1016/0022-2836(86)90225-1. [DOI] [PubMed] [Google Scholar]
- Liu S. L., Hessel A., Sanderson K. E. Genomic mapping with I-Ceu I, an intron-encoded endonuclease specific for genes for ribosomal RNA, in Salmonella spp., Escherichia coli, and other bacteria. Proc Natl Acad Sci U S A. 1993 Jul 15;90(14):6874–6878. doi: 10.1073/pnas.90.14.6874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S. L., Hessel A., Sanderson K. E. The XbaI-BlnI-CeuI genomic cleavage map of Salmonella typhimurium LT2 determined by double digestion, end labelling, and pulsed-field gel electrophoresis. J Bacteriol. 1993 Jul;175(13):4104–4120. doi: 10.1128/jb.175.13.4104-4120.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S. L., Sanderson K. E. A physical map of the Salmonella typhimurium LT2 genome made by using XbaI analysis. J Bacteriol. 1992 Mar;174(5):1662–1672. doi: 10.1128/jb.174.5.1662-1672.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macnab R. M. Genetics and biogenesis of bacterial flagella. Annu Rev Genet. 1992;26:131–158. doi: 10.1146/annurev.ge.26.120192.001023. [DOI] [PubMed] [Google Scholar]
- O'Reilly C., Black G. W., Laffey R., McConnell D. J. Molecular analysis of an IS200 insertion in the gpt gene of Salmonella typhimurium LT2. J Bacteriol. 1990 Nov;172(11):6599–6601. doi: 10.1128/jb.172.11.6599-6601.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnishi K., Kutsukake K., Suzuki H., Iino T. Gene fliA encodes an alternative sigma factor specific for flagellar operons in Salmonella typhimurium. Mol Gen Genet. 1990 Apr;221(2):139–147. doi: 10.1007/BF00261713. [DOI] [PubMed] [Google Scholar]
- Sanderson K. E. Genetic relatedness in the family Enterobacteriaceae. Annu Rev Microbiol. 1976;30:327–349. doi: 10.1146/annurev.mi.30.100176.001551. [DOI] [PubMed] [Google Scholar]
- Sanderson K. E., Roth J. R. Linkage map of Salmonella typhimurium, edition VII. Microbiol Rev. 1988 Dec;52(4):485–532. doi: 10.1128/mr.52.4.485-532.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderson K. E., Stocker B. A. Gene rfaH, which affects lipopolysaccharide core structure in Salmonella typhimurium, is required also for expression of F-factor functions. J Bacteriol. 1981 May;146(2):535–541. doi: 10.1128/jb.146.2.535-541.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro J. A. Mutations caused by the insertion of genetic material into the galactose operon of Escherichia coli. J Mol Biol. 1969 Feb 28;40(1):93–105. doi: 10.1016/0022-2836(69)90298-8. [DOI] [PubMed] [Google Scholar]
- Stanley J., Baquar N., Threlfall E. J. Genotypes and phylogenetic relationships of Salmonella typhimurium are defined by molecular fingerprinting of IS200 and 16S rrn loci. J Gen Microbiol. 1993 Jun;139(Pt 6):1133–1140. doi: 10.1099/00221287-139-6-1133. [DOI] [PubMed] [Google Scholar]
- Stanley J., Burnens A., Powell N., Chowdry N., Jones C. The insertion sequence IS200 fingerprints chromosomal genotypes and epidemiological relationships in Salmonella heidelberg. J Gen Microbiol. 1992 Nov;138(11):2329–2336. doi: 10.1099/00221287-138-11-2329. [DOI] [PubMed] [Google Scholar]
- Stanley J., Chowdry N., Powell N., Threlfall E. J. Chromosomal genotypes (evolutionary lines) of Salmonella berta. FEMS Microbiol Lett. 1992 Aug 15;74(2-3):247–252. doi: 10.1016/0378-1097(92)90437-s. [DOI] [PubMed] [Google Scholar]
- Stanley J., Goldsworthy M., Threlfall E. J. Molecular phylogenetic typing of pandemic isolates of Salmonella enteritidis. FEMS Microbiol Lett. 1992 Jan 1;69(2):153–160. doi: 10.1016/0378-1097(92)90620-4. [DOI] [PubMed] [Google Scholar]
- Wong K. K., McClelland M. A BlnI restriction map of the Salmonella typhimurium LT2 genome. J Bacteriol. 1992 Mar;174(5):1656–1661. doi: 10.1128/jb.174.5.1656-1661.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T. T. A model for three-point analysis of random general transduction. Genetics. 1966 Aug;54(2):405–410. doi: 10.1093/genetics/54.2.405. [DOI] [PMC free article] [PubMed] [Google Scholar]




