Abstract
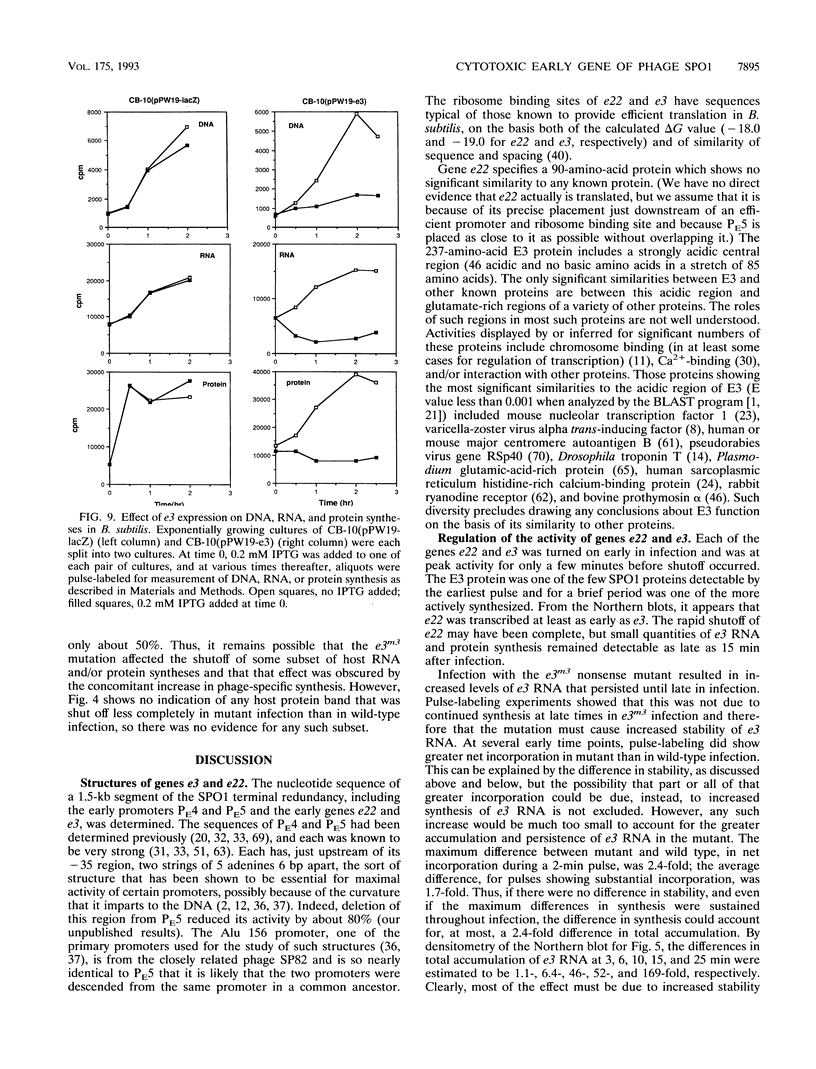
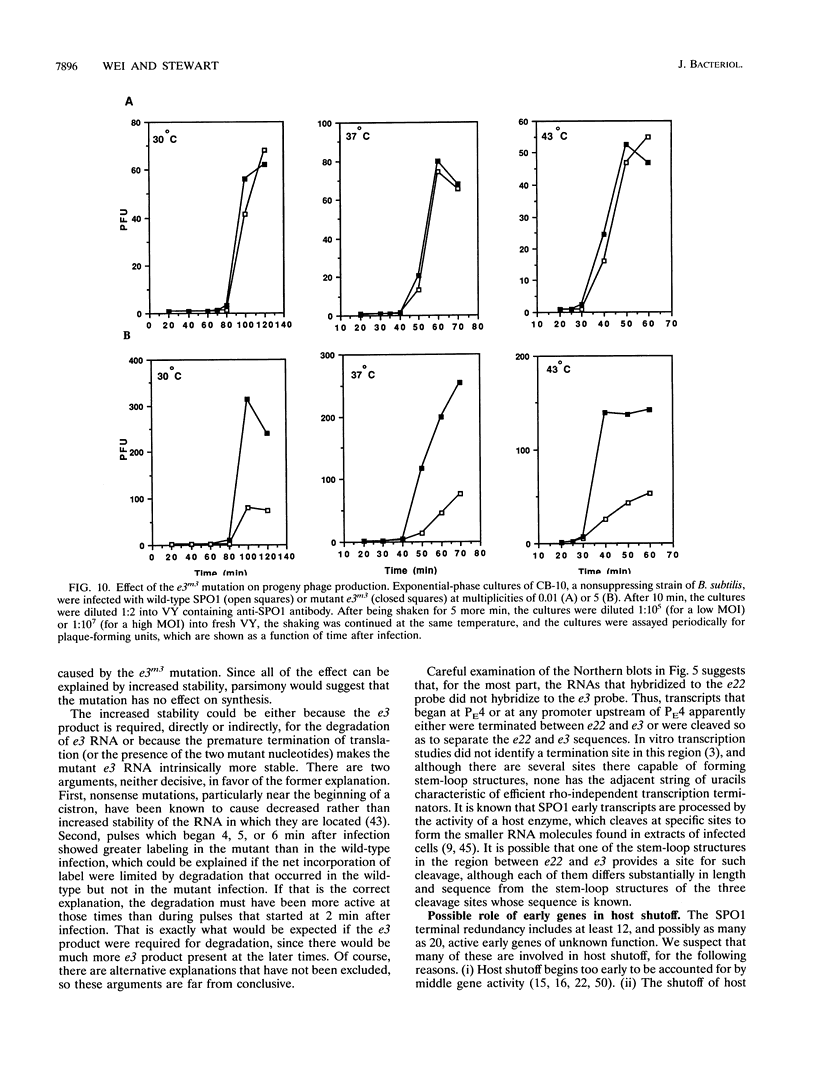
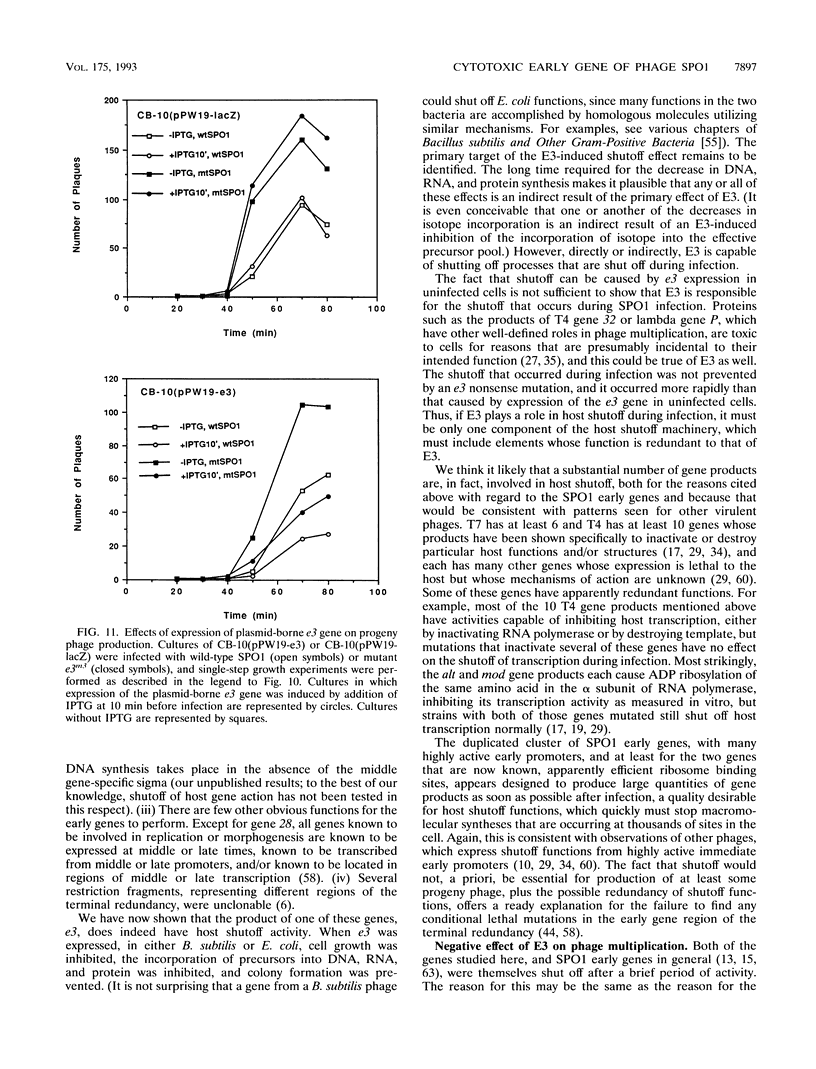
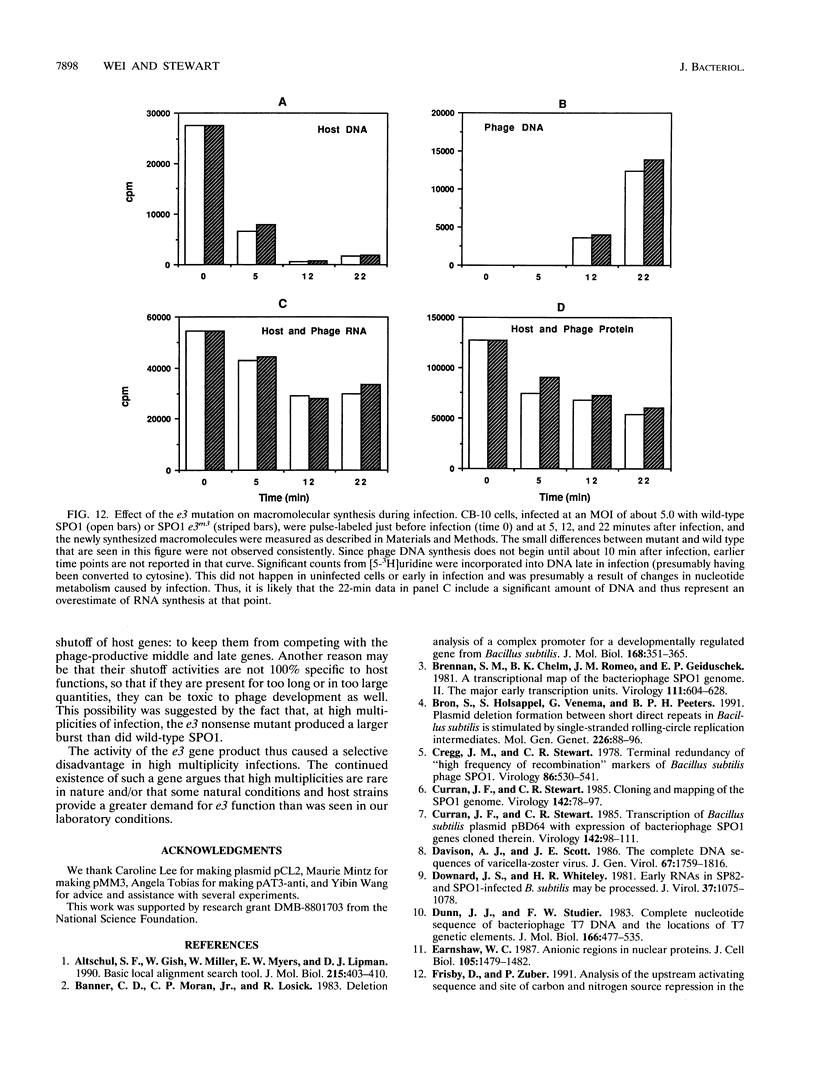
Some of the early genes of Bacillus subtilis bacteriophage SPO1 were hypothesized to function in the shutoff of host biosyntheses. Two of these genes, e3 and e22, were cloned and sequenced. E22 showed no similarity to any known protein, while E3, a highly acidic protein, showed significant similarity only to other similarly acidic proteins. Each gene was immediately downstream of a very active early promoter. Each was expressed actively during the first few minutes of infection and was then rapidly shut off and its RNA rapidly degraded. An e3 nonsense mutation severely retarded the degradation of e3 RNA. Expression of a plasmid-borne e3 gene, in either B. subtilis or Escherichia coli, resulted in the inhibition of host DNA, RNA, and protein syntheses and prevented colony formation. However, the e3 nonsense mutation caused no measurable decrease in either burst size or host shutoff during infection and, in fact, caused an increased burst size at high multiplicities of infection. We suggest that e3 is one of several genes involved in host shutoff, that its function is dispensable both for host shutoff and for phage multiplication, and that its shutoff function is not entirely specific to host activities.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. Basic local alignment search tool. J Mol Biol. 1990 Oct 5;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Banner C. D., Moran C. P., Jr, Losick R. Deletion analysis of a complex promoter for a developmentally regulated gene from Bacillus subtilis. J Mol Biol. 1983 Aug 5;168(2):351–365. doi: 10.1016/s0022-2836(83)80023-0. [DOI] [PubMed] [Google Scholar]
- Bron S., Holsappel S., Venema G., Peeters B. P. Plasmid deletion formation between short direct repeats in Bacillus subtilis is stimulated by single-stranded rolling-circle replication intermediates. Mol Gen Genet. 1991 Apr;226(1-2):88–96. doi: 10.1007/BF00273591. [DOI] [PubMed] [Google Scholar]
- Chelm B. K., Romeo J. M., Brennan S. M., Geiduschek E. P. A transcriptional map of the bacteriophage SPO1 genome. III. A region of early and middle promoters (the gene 28 region). Virology. 1981 Jul 30;112(2):572–588. doi: 10.1016/0042-6822(81)90303-2. [DOI] [PubMed] [Google Scholar]
- Cregg J. M., Stewart C. R. Terminal redundancy of "high frequency of recombination" markers of Bacillus subtilis phage SPO1. Virology. 1978 May 15;86(2):530–541. doi: 10.1016/0042-6822(78)90091-0. [DOI] [PubMed] [Google Scholar]
- Curran J. F., Stewart C. R. Cloning and mapping of the SPO1 genome. Virology. 1985 Apr 15;142(1):78–97. doi: 10.1016/0042-6822(85)90424-6. [DOI] [PubMed] [Google Scholar]
- Curran J. F., Stewart C. R. Transcription of Bacillis subtilis plasmid pBD64 and expression of bacteriophage SPO1 genes cloned therein. Virology. 1985 Apr 15;142(1):98–111. doi: 10.1016/0042-6822(85)90425-8. [DOI] [PubMed] [Google Scholar]
- Davison A. J., Scott J. E. The complete DNA sequence of varicella-zoster virus. J Gen Virol. 1986 Sep;67(Pt 9):1759–1816. doi: 10.1099/0022-1317-67-9-1759. [DOI] [PubMed] [Google Scholar]
- Downard J. S., Whiteley H. R. Early RNAs in SP82- and SP01-infected Bacillus subtilis may be processed. J Virol. 1981 Mar;37(3):1075–1078. doi: 10.1128/jvi.37.3.1075-1078.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn J. J., Studier F. W. Complete nucleotide sequence of bacteriophage T7 DNA and the locations of T7 genetic elements. J Mol Biol. 1983 Jun 5;166(4):477–535. doi: 10.1016/s0022-2836(83)80282-4. [DOI] [PubMed] [Google Scholar]
- Earnshaw W. C. Anionic regions in nuclear proteins. J Cell Biol. 1987 Oct;105(4):1479–1482. doi: 10.1083/jcb.105.4.1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frisby D., Zuber P. Analysis of the upstream activating sequence and site of carbon and nitrogen source repression in the promoter of an early-induced sporulation gene of Bacillus subtilis. J Bacteriol. 1991 Dec;173(23):7557–7564. doi: 10.1128/jb.173.23.7557-7564.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita D. J., Ohlsson-Wilhelm B. M., Geiduschek E. P. Transcription during bacteriophage SPO1 development: mutations affecting the program of viral transcription. J Mol Biol. 1971 Apr 28;57(2):301–317. doi: 10.1016/0022-2836(71)90348-2. [DOI] [PubMed] [Google Scholar]
- Fyrberg E., Fyrberg C. C., Beall C., Saville D. L. Drosophila melanogaster troponin-T mutations engender three distinct syndromes of myofibrillar abnormalities. J Mol Biol. 1990 Dec 5;216(3):657–675. doi: 10.1016/0022-2836(90)90390-8. [DOI] [PubMed] [Google Scholar]
- Gage L. P., Geiduschek E. P. RNA synthesis during bacteriophage SPO1 development: six classes of SPO1 RNA. J Mol Biol. 1971 Apr 28;57(2):279–297. doi: 10.1016/0022-2836(71)90346-9. [DOI] [PubMed] [Google Scholar]
- Gage L. P., Geiduschek E. P. RNA synthesis during bacteriphage SPO1 development. II. Some modulations and prerequisites of the transcription program. Virology. 1971 Apr;44(1):200–210. doi: 10.1016/0042-6822(71)90165-6. [DOI] [PubMed] [Google Scholar]
- Glassberg J., Franck M., Stewart C. R. Initiation and termination mutants of Bacillus subtilis bacteriophage SPO1. J Virol. 1977 Jan;21(1):147–152. doi: 10.1128/jvi.21.1.147-152.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goff C. G., Setzer J. ADP ribosylation of Escherichia coli RNA polymerase is nonessential for bacteriophage T4 development. J Virol. 1980 Jan;33(1):547–549. doi: 10.1128/jvi.33.1.547-549.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene J. R., Morrissey L. M., Foster L. M., Geiduschek E. P. DNA binding by the bacteriophage SPO1-encoded type II DNA-binding protein, transcription factor 1. Formation of nested complexes at a selective binding site. J Biol Chem. 1986 Sep 25;261(27):12820–12827. [PubMed] [Google Scholar]
- Heintz N., Shub D. A. Transcriptional regulation of bacteriophage SPO1 protein synthesis in vivo and in vitro. J Virol. 1982 Jun;42(3):951–962. doi: 10.1128/jvi.42.3.951-962.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hisatake K., Nishimura T., Maeda Y., Hanada K., Song C. Z., Muramatsu M. Cloning and structural analysis of cDNA and the gene for mouse transcription factor UBF. Nucleic Acids Res. 1991 Sep 11;19(17):4631–4637. doi: 10.1093/nar/19.17.4631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann S. L., Topham M., Hsieh C. L., Francke U. cDNA and genomic cloning of HRC, a human sarcoplasmic reticulum protein, and localization of the gene to human chromosome 19 and mouse chromosome 7. Genomics. 1991 Apr;9(4):656–669. doi: 10.1016/0888-7543(91)90359-m. [DOI] [PubMed] [Google Scholar]
- Horinouchi S., Weisblum B. Nucleotide sequence and functional map of pC194, a plasmid that specifies inducible chloramphenicol resistance. J Bacteriol. 1982 May;150(2):815–825. doi: 10.1128/jb.150.2.815-825.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jannière L., Bruand C., Ehrlich S. D. Structurally stable Bacillus subtilis cloning vectors. Gene. 1990 Mar 1;87(1):53–61. doi: 10.1016/0378-1119(90)90495-d. [DOI] [PubMed] [Google Scholar]
- Krisch H. M., Selzer G. B. Construction and properties of a recombinant plasmid containing gene 32 of bacteriophage T4D. J Mol Biol. 1981 May 25;148(3):199–218. doi: 10.1016/0022-2836(81)90535-0. [DOI] [PubMed] [Google Scholar]
- Kunkel T. A., Roberts J. D., Zakour R. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- Leberer E., Charuk J. H., Green N. M., MacLennan D. H. Molecular cloning and expression of cDNA encoding a lumenal calcium binding glycoprotein from sarcoplasmic reticulum. Proc Natl Acad Sci U S A. 1989 Aug;86(16):6047–6051. doi: 10.1073/pnas.86.16.6047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee G., Hannett N. M., Korman A., Pero J. Transcription of cloned DNA from Bacillus subtilis phage SP01. Requirement for hydroxymethyluracil-containing DNA by phage-modified RNA polymerase. J Mol Biol. 1980 May 25;139(3):407–422. doi: 10.1016/0022-2836(80)90138-2. [DOI] [PubMed] [Google Scholar]
- Lee G., Pero J. Conserved nucleotide sequences in temporally controlled bacteriophage promoters. J Mol Biol. 1981 Oct 25;152(2):247–265. doi: 10.1016/0022-2836(81)90242-4. [DOI] [PubMed] [Google Scholar]
- Lee G., Talkington C., Pero J. Nucleotide sequence of a promoter recognized by Bacillus subtilis RNA polymerase. Mol Gen Genet. 1980;180(1):57–65. doi: 10.1007/BF00267352. [DOI] [PubMed] [Google Scholar]
- Liu Q., Richardson C. C. Gene 5.5 protein of bacteriophage T7 inhibits the nucleoid protein H-NS of Escherichia coli. Proc Natl Acad Sci U S A. 1993 Mar 1;90(5):1761–1765. doi: 10.1073/pnas.90.5.1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiti S., Mukhopadhyay M., Mandal N. C. Bacteriophage lambda P gene shows host killing which is not dependent on lambda DNA replication. Virology. 1991 May;182(1):324–335. doi: 10.1016/0042-6822(91)90676-3. [DOI] [PubMed] [Google Scholar]
- McAllister C. F., Achberger E. C. Effect of polyadenine-containing curved DNA on promoter utilization in Bacillus subtilis. J Biol Chem. 1988 Aug 25;263(24):11743–11749. [PubMed] [Google Scholar]
- McAllister C. F., Achberger E. C. Rotational orientation of upstream curved DNA affects promoter function in Bacillus subtilis. J Biol Chem. 1989 Jun 25;264(18):10451–10456. [PubMed] [Google Scholar]
- McKenzie T., Hoshino T., Tanaka T., Sueoka N. Correction. A revision of the nucleotide sequence and functional map of pUB110. Plasmid. 1987 Jan;17(1):83–85. doi: 10.1016/0147-619x(87)90015-1. [DOI] [PubMed] [Google Scholar]
- McLaughlin J. R., Murray C. L., Rabinowitz J. C. Unique features in the ribosome binding site sequence of the gram-positive Staphylococcus aureus beta-lactamase gene. J Biol Chem. 1981 Nov 10;256(21):11283–11291. [PubMed] [Google Scholar]
- Mulbry W. W., Ambulos N. P., Jr, Lovett P. S. Bacillus subtilis mutant allele sup-3 causes lysine insertion at ochre codons: use of sup-3 in studies of translational attenuation. J Bacteriol. 1989 Oct;171(10):5322–5324. doi: 10.1128/jb.171.10.5322-5324.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray C. L., Rabinowitz J. C. Nucleotide sequences of transcription and translation initiation regions in Bacillus phage phi 29 early genes. J Biol Chem. 1982 Jan 25;257(2):1053–1062. [PubMed] [Google Scholar]
- Nilsson G., Belasco J. G., Cohen S. N., von Gabain A. Effect of premature termination of translation on mRNA stability depends on the site of ribosome release. Proc Natl Acad Sci U S A. 1987 Jul;84(14):4890–4894. doi: 10.1073/pnas.84.14.4890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okubo S., Yanagida T., Fujita D. J., Olsson-Wilhelm B. M. The genetics of bacteriophage SPO1. Biken J. 1972 Jun;15(2):81–97. [PubMed] [Google Scholar]
- Panganiban A. T., Whiteley H. R. Bacillus subtilis RNAase III cleavage sites in phage SP82 early mRNA. Cell. 1983 Jul;33(3):907–913. doi: 10.1016/0092-8674(83)90033-8. [DOI] [PubMed] [Google Scholar]
- Panneerselvam C., Wellner D., Horecker B. L. The amino acid sequence of bovine thymus prothymosin alpha. Arch Biochem Biophys. 1988 Sep;265(2):454–457. doi: 10.1016/0003-9861(88)90149-x. [DOI] [PubMed] [Google Scholar]
- Perkus M. E., Shub D. A. Mapping the genes in the terminal redundancy of bacteriophage SPO1 with restriction endonucleases. J Virol. 1985 Oct;56(1):40–48. doi: 10.1128/jvi.56.1.40-48.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pero J., Hannett N. M., Talkington C. Restriction cleavage map of SP01 DNA: general location of early, middle, and late genes. J Virol. 1979 Jul;31(1):156–171. doi: 10.1128/jvi.31.1.156-171.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeve J. N., Mertens G., Amann E. Early development of bacteriophages SP01 and SP82G in minicells of Bacillus subtilis. J Mol Biol. 1978 Apr 5;120(2):183–207. doi: 10.1016/0022-2836(78)90064-5. [DOI] [PubMed] [Google Scholar]
- Sayre M. H., Geiduschek E. P. TF1, the bacteriophage SPO1-encoded type II DNA-binding protein, is essential for viral multiplication. J Virol. 1988 Sep;62(9):3455–3462. doi: 10.1128/jvi.62.9.3455-3462.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimotsu H., Henner D. J. Construction of a single-copy integration vector and its use in analysis of regulation of the trp operon of Bacillus subtilis. Gene. 1986;43(1-2):85–94. doi: 10.1016/0378-1119(86)90011-9. [DOI] [PubMed] [Google Scholar]
- Spizizen J. TRANSFORMATION OF BIOCHEMICALLY DEFICIENT STRAINS OF BACILLUS SUBTILIS BY DEOXYRIBONUCLEATE. Proc Natl Acad Sci U S A. 1958 Oct 15;44(10):1072–1078. doi: 10.1073/pnas.44.10.1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart C. R., Cater M., Click B. Lysis of Bacillus subtilis by bacteriophage SP82 in the absence of DNA synthesis. Virology. 1971 Nov;46(2):327–336. doi: 10.1016/0042-6822(71)90034-1. [DOI] [PubMed] [Google Scholar]
- Stewart C. R. Physical heterogeneity among Bacillus subtilis deoxyribonucleic acid molecules carrying particular genetic markers. J Bacteriol. 1969 Jun;98(3):1239–1247. doi: 10.1128/jb.98.3.1239-1247.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studier F. W. Use of bacteriophage T7 lysozyme to improve an inducible T7 expression system. J Mol Biol. 1991 May 5;219(1):37–44. doi: 10.1016/0022-2836(91)90855-z. [DOI] [PubMed] [Google Scholar]
- Sullivan K. F., Glass C. A. CENP-B is a highly conserved mammalian centromere protein with homology to the helix-loop-helix family of proteins. Chromosoma. 1991 Jul;100(6):360–370. doi: 10.1007/BF00337514. [DOI] [PubMed] [Google Scholar]
- Takeshima H., Nishimura S., Matsumoto T., Ishida H., Kangawa K., Minamino N., Matsuo H., Ueda M., Hanaoka M., Hirose T. Primary structure and expression from complementary DNA of skeletal muscle ryanodine receptor. Nature. 1989 Jun 8;339(6224):439–445. doi: 10.1038/339439a0. [DOI] [PubMed] [Google Scholar]
- Talkington C., Pero J. Restriction fragment analysis of the temporal program of bacteriophage SPO1 transcription and its control by phage-modified RNA polymerases. Virology. 1977 Dec;83(2):365–379. doi: 10.1016/0042-6822(77)90181-7. [DOI] [PubMed] [Google Scholar]
- Tinoco I., Jr, Borer P. N., Dengler B., Levin M. D., Uhlenbeck O. C., Crothers D. M., Bralla J. Improved estimation of secondary structure in ribonucleic acids. Nat New Biol. 1973 Nov 14;246(150):40–41. doi: 10.1038/newbio246040a0. [DOI] [PubMed] [Google Scholar]
- Triglia T., Stahl H. D., Crewther P. E., Silva A., Anders R. F., Kemp D. J. Structure of a Plasmodium falciparum gene that encodes a glutamic acid-rich protein (GARP). Mol Biochem Parasitol. 1988 Nov;31(2):199–201. doi: 10.1016/0166-6851(88)90170-3. [DOI] [PubMed] [Google Scholar]
- Vieira J., Messing J. Production of single-stranded plasmid DNA. Methods Enzymol. 1987;153:3–11. doi: 10.1016/0076-6879(87)53044-0. [DOI] [PubMed] [Google Scholar]
- Wang T., Okano Y., Eisensmith R. C., Fekete G., Schuler D., Berencsi G., Nasz I., Woo S. L. Molecular genetics of PKU in eastern Europe: a nonsense mutation associated with haplotype 4 of the phenylalanine hydroxylase gene. Somat Cell Mol Genet. 1990 Jan;16(1):85–90. doi: 10.1007/BF01650483. [DOI] [PubMed] [Google Scholar]
- Wilson D. L., Gage L. P. Certain aspects of SPO1 development. J Mol Biol. 1971 Apr 28;57(2):297–300. doi: 10.1016/0022-2836(71)90347-0. [DOI] [PubMed] [Google Scholar]
- Yansura D. G., Henner D. J. Use of the Escherichia coli lac repressor and operator to control gene expression in Bacillus subtilis. Proc Natl Acad Sci U S A. 1984 Jan;81(2):439–443. doi: 10.1073/pnas.81.2.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G., Leader D. P. The structure of the pseudorabies virus genome at the end of the inverted repeat sequences proximal to the junction with the short unique region. J Gen Virol. 1990 Oct;71(Pt 10):2433–2441. doi: 10.1099/0022-1317-71-10-2433. [DOI] [PubMed] [Google Scholar]
- Zuber P., Losick R. Use of a lacZ fusion to study the role of the spoO genes of Bacillus subtilis in developmental regulation. Cell. 1983 Nov;35(1):275–283. doi: 10.1016/0092-8674(83)90230-1. [DOI] [PubMed] [Google Scholar]