Abstract
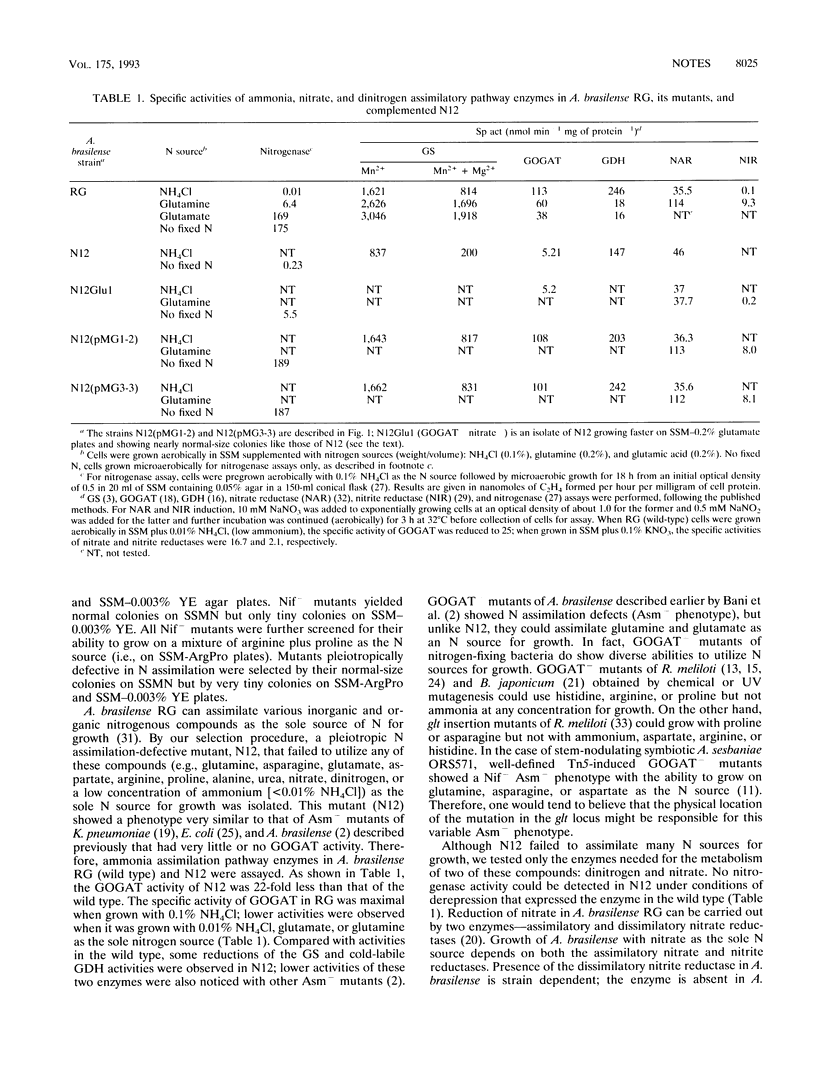
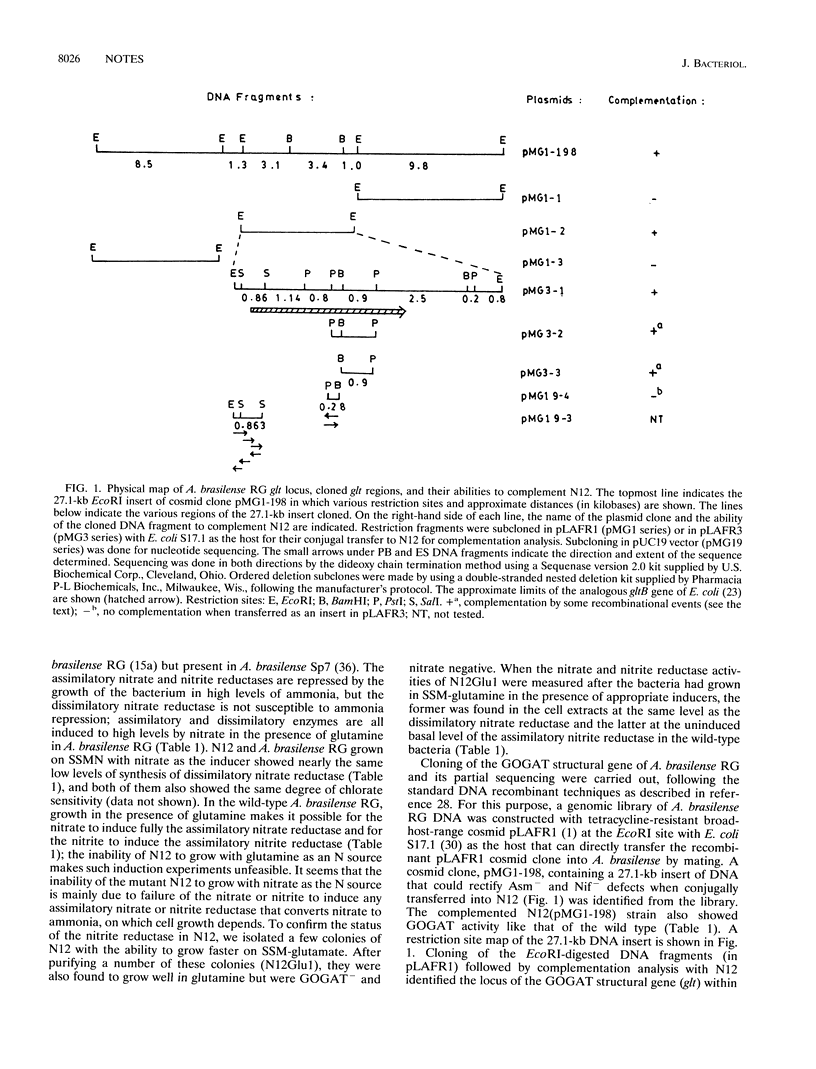
An Azospirillum brasilense mutant (N12) pleiotropically defective in the assimilation of nitrogenous compounds (Asm-) was isolated and found lacking in the glutamate synthase (GOGAT-). The glt (GOGAT) locus of A. brasilense was identified by isolating a broad-host-range pLAFR1 cosmid clone from a gene library of the bacterium that rectified Asm- and GOGAT- defects (full recovery of activities of the nitrogenase, the assimilatory nitrate and nitrite reductases, and the glutamate synthase). A 7.5-kb EcoRI fragment of the cosmid clone that also complemented N12 was partially sequenced to identify the open reading frame for the alpha-subunit of GOGAT. The amino acid sequences deduced from the partial nucleotide sequences of the glt locus of A. brasilense showed considerable homology with that of the alpha-subunit of GOGAT coded by the gltB gene of Escherichia coli. The genetic lesion of N12 was found within the gltB gene of A. brasilense. The gltB promoter of A. brasilense showed the presence of a consensus sigma-70-like recognition site (as in E. coli) in addition to potential NtrA-RNA polymerase, IHF, and NifA binding sites.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bender R. A., Janssen K. A., Resnick A. D., Blumenberg M., Foor F., Magasanik B. Biochemical parameters of glutamine synthetase from Klebsiella aerogenes. J Bacteriol. 1977 Feb;129(2):1001–1009. doi: 10.1128/jb.129.2.1001-1009.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozouklian H., Fogher C., Elmerich C. Cloning and characterization of the glnA gene of Azospirillum brasilense Sp7. Ann Inst Pasteur Microbiol. 1986 Jul-Aug;137B(1):3–18. doi: 10.1016/s0769-2609(86)80089-8. [DOI] [PubMed] [Google Scholar]
- Castaño I., Bastarrachea F., Covarrubias A. A. gltBDF operon of Escherichia coli. J Bacteriol. 1988 Feb;170(2):821–827. doi: 10.1128/jb.170.2.821-827.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collado-Vides J., Magasanik B., Gralla J. D. Control site location and transcriptional regulation in Escherichia coli. Microbiol Rev. 1991 Sep;55(3):371–394. doi: 10.1128/mr.55.3.371-394.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernsting B. R., Atkinson M. R., Ninfa A. J., Matthews R. G. Characterization of the regulon controlled by the leucine-responsive regulatory protein in Escherichia coli. J Bacteriol. 1992 Feb;174(4):1109–1118. doi: 10.1128/jb.174.4.1109-1118.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman A. M., Long S. R., Brown S. E., Buikema W. J., Ausubel F. M. Construction of a broad host range cosmid cloning vector and its use in the genetic analysis of Rhizobium mutants. Gene. 1982 Jun;18(3):289–296. doi: 10.1016/0378-1119(82)90167-6. [DOI] [PubMed] [Google Scholar]
- Gosset G., Merino E., Recillas F., Oliver G., Becerril B., Bolivar F. Amino acid sequence analysis of the glutamate synthase enzyme from Escherichia coli K-12. Protein Seq Data Anal. 1989;2(1):9–16. [PubMed] [Google Scholar]
- Hoover T. R., Santero E., Porter S., Kustu S. The integration host factor stimulates interaction of RNA polymerase with NIFA, the transcriptional activator for nitrogen fixation operons. Cell. 1990 Oct 5;63(1):11–22. doi: 10.1016/0092-8674(90)90284-l. [DOI] [PubMed] [Google Scholar]
- Kuczius T., Eitinger T., D'Ari R., Castorph H., Kleiner D. The gltF gene of Klebsiella pneumoniae: cloning and initial characterization. Mol Gen Genet. 1991 Oct;229(3):479–482. doi: 10.1007/BF00267472. [DOI] [PubMed] [Google Scholar]
- Lewis T. A., Gonzalez R., Botsford J. L. Rhizobium meliloti glutamate synthase: cloning and initial characterization of the glt locus. J Bacteriol. 1990 May;172(5):2413–2420. doi: 10.1128/jb.172.5.2413-2420.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis T. A., Gonzalez R., Botsford J. L. Rhizobium meliloti glutamate synthase: cloning and initial characterization of the glt locus. J Bacteriol. 1990 May;172(5):2413–2420. doi: 10.1128/jb.172.5.2413-2420.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maulik P., Ghosh S. NADPH/NADH-dependent cold-labile glutamate dehydrogenase in Azospirillum brasilense. Purification and properties. Eur J Biochem. 1986 Mar 17;155(3):595–602. doi: 10.1111/j.1432-1033.1986.tb09530.x. [DOI] [PubMed] [Google Scholar]
- Miller R. E., Stadtman E. R. Glutamate synthase from Escherichia coli. An iron-sulfide flavoprotein. J Biol Chem. 1972 Nov 25;247(22):7407–7419. [PubMed] [Google Scholar]
- Nagatani H., Shimizu M., Valentine R. C. The mechanism of ammonia assimilation in nitrogen fixing Bacteria. Arch Mikrobiol. 1971;79(2):164–175. doi: 10.1007/BF00424923. [DOI] [PubMed] [Google Scholar]
- Neyra C. A., Van Berkum P. Nitrate reduction nitrogenase activity in Spirillum lipoferum1. Can J Microbiol. 1977 Mar;23(3):306–310. doi: 10.1139/m77-045. [DOI] [PubMed] [Google Scholar]
- Oliver G., Gosset G., Sanchez-Pescador R., Lozoya E., Ku L. M., Flores N., Becerril B., Valle F., Bolivar F. Determination of the nucleotide sequence for the glutamate synthase structural genes of Escherichia coli K-12. Gene. 1987;60(1):1–11. doi: 10.1016/0378-1119(87)90207-1. [DOI] [PubMed] [Google Scholar]
- Osburne M. S., Signer E. R. Ammonium assimilation in Rhizobium meliloti. J Bacteriol. 1980 Sep;143(3):1234–1240. doi: 10.1128/jb.143.3.1234-1240.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pahel G., Zelenetz A. D., Tyler B. M. gltB gene and regulation of nitrogen metabolism by glutamine synthetase in Escherichia coli. J Bacteriol. 1978 Jan;133(1):139–148. doi: 10.1128/jb.133.1.139-148.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratti S., Curti B., Zanetti G., Galli E. Purification and characterization of glutamate synthase from Azospirillum brasilense. J Bacteriol. 1985 Aug;163(2):724–729. doi: 10.1128/jb.163.2.724-729.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauth S., Ghosh S. Effect of dimethylsulfoxide on derepression of nitrogenase in Spirillum lipoferum. FEBS Lett. 1981 Apr 6;126(1):77–80. doi: 10.1016/0014-5793(81)81037-x. [DOI] [PubMed] [Google Scholar]
- Santero E., Luque F., Medina J. R., Tortolero M. Isolation of ntrA-like mutants of Azotobacter vinelandii. J Bacteriol. 1986 May;166(2):541–544. doi: 10.1128/jb.166.2.541-544.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toukdarian A., Kennedy C. Regulation of nitrogen metabolism in Azotobacter vinelandii: isolation of ntr and glnA genes and construction of ntr mutants. EMBO J. 1986 Feb;5(2):399–407. doi: 10.1002/j.1460-2075.1986.tb04225.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanoni M. A., Negri A., Zanetti G., Ronchi S., Curti B. Structural studies on the subunits of glutamate synthase from Azospirillum brasilense. Biochim Biophys Acta. 1990 Jul 6;1039(3):374–377. doi: 10.1016/0167-4838(90)90273-i. [DOI] [PubMed] [Google Scholar]
- Westby C. A., Enderlin C. S., Steinberg N. A., Joseph C. M., Meeks J. C. Assimilation of 13NH4+ by Azospirillum brasilense grown under nitrogen limitation and excess. J Bacteriol. 1987 Sep;169(9):4211–4214. doi: 10.1128/jb.169.9.4211-4214.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Zamaroczy M., Delorme F., Elmerich C. Regulation of transcription and promoter mapping of the structural genes for nitrogenase (nifHDK) of Azospirillum brasilense Sp7. Mol Gen Genet. 1989 Dec;220(1):88–94. doi: 10.1007/BF00260861. [DOI] [PubMed] [Google Scholar]



