Abstract
Plectin is a major component of the cytoskeleton and links the intermediate filament system to hemidesmosomes by binding to the integrin β4 subunit. Previously, a binding site for β4 was mapped on the actin-binding domain (ABD) of plectin and binding of β4 and F-actin to plectin was shown to be mutually exclusive. Here we show that only the ABDs of plectin and dystonin bind to β4, whereas those of other actin-binding proteins do not. Mutations of the ABD of plectin-1C show that Q131, R138, and N149 are critical for tight binding of the ABD to β4. These residues form a small cavity, occupied by a well-ordered water molecule in the crystal structure. The β4 binding pocket partly overlaps with the actin-binding sequence 2 (ABS2), previously shown to be essential for actin binding. Therefore, steric interference may render binding of β4 and F-actin to plectin mutually exclusive. Finally, we provide evidence indicating that the residues preceding the ABD in plectin-1A and -1C, although unable to mediate binding to β4 themselves, modulate the binding activity of the ABD for β4. These studies demonstrate the unique property of the plectin-ABD to bind to both F-actin and β4, and explain why several other ABD-containing proteins that are expressed in basal keratinocytes are not recruited into hemidesmosomes.
INTRODUCTION
Anchoring of cells to the basement membrane is crucial for the function and integrity of epithelial tissues. Hemidesmosomes are protein complexes that mediate stable anchoring by providing a tight link between the intracellular intermediate filament system and the extracellular matrix. They are assembled at the basal side of basal epithelial cells in (pseudo-) stratified and some complex epithelia. Hemidesmosomes consist of at least five distinct proteins. Three of these are transmembrane proteins: the integrin α6β4 (Stepp et al., 1990; Sonnenberg et al., 1991; Jones et al., 1991), the bullous pemphigoid antigen 180 (BP180; Giudice et al., 1992), and the tetraspanin CD151 (Sterk et al., 2000). The two cytoplasmic proteins, BP230 and plectin, that are localized in the hemidesmosomal plaque play a major role in linking the intermediate filament system to the hemidesmosome (Borradori and Sonnenberg, 1996; Green and Jones, 1996; Burgeson and Christiano, 1997).
The interaction of α6β4 with plectin is essential for establishing the link between the extracellular matrix and the intermediate filament system. Inactivation of the genes for either α6 or β4 in humans results in a severe and fatal skin blistering disease, called pyloric atresia associated with junctional epidermolysis bullosa (PA-JEB; Vidal et al., 1995; Ruzzi et al., 1997). A similar phenotype is observed in genetically modified mice that lack either α6 or β4 (van der Neut et al., 1996, Dowling et al., 1996, Georges-Labouesse et al., 1996). Similarly, the loss of or a reduced expression of plectin leads to a blistering disorder, called epidermolysis bullosa simplex associated with muscular dystrophy (MD-EBS; Gache et al., 1996; McLean et al., 1996; Smith et al., 1996; Andrä et al., 1997). These examples of both human patients and mice show the importance of hemidesmosomes for the stable adhesion of the epidermis to the dermis as well as for tissue integrity. Most of the mutations identified in PA-JEB patients are nonsense mutations or mutations at splice sites that result in the early termination of translation of the β4 protein. Missense mutations, resulting in the substitution of a single amino acid have also been described. Two of these point mutations (R1225H and R1281W) have been disclosed in patients with a nonlethal form of epidermolysis bullosa (EB; Pulkkinen et al., 1998; Nakano et al., 2001), and recently these mutations were shown to result in the inability of β4 to recruit plectin into hemidesmosomes (Koster et al., 2001).
Plectin is a widely expressed cytoskeletal linker protein of >500 kDa that interacts with actin, intermediate filaments and microtubules (for a review see Steinbock and Wiche, 1999). It belongs to the plakin family of proteins, the members of which share a similar multi-domain structure: a long central coiled-coil rod domain, flanked by N- and C-terminal globular domains. The central rod domain mediates dimerization and/or multimerization of plectin (Foisner and Wiche, 1987; Wiche, 1998). The C-terminal domain contains a binding site for intermediate filament proteins. The N-terminal domain contains a highly conserved actin-binding domain (ABD) of the β-spectrin type (McLean et al., 1996). This type of ABD is found in many actin-binding proteins, including dystonin, α-actinin, utrophin, filamin, and dystrophin, and consists of a pair of calponin homology (CH) domains (for reviews, see Hartwig, 1994; Gimona et al., 2002). Plectin is encoded by the PLEC1 gene, which is a large and complex gene containing several alternative first exons. Each of these first exons can be spliced into a common exon 2, encoding the start of the ABD (encoded by exons 2–8), thus generating a variety of plectin variants. The resulting plectin splice variants have characteristic tissue expression and actin binding properties (Fuchs et al., 1999).
Previously, we showed that plectin binds to the β4 subunit of the integrin α6β4 via its N-terminal ABD and that this interaction prevents the association of the ABD with F-actin (Geerts et al., 1999). This suggests that the binding sites on the ABD for F-actin and β4 overlap or even are identical. In the calponin-type ABD three regions are essential for actin binding, i.e., the actin-binding sequences 1–3 (ABS1–3; Bresnick et al., 1990; Levine et al., 1992). No specific sequence in the ABD, which interacts with β4, has yet been identified. Furthermore, the influence on β4 binding of the variable sequences preceding the ABD, which vary from 5–180 amino acids in length and share no sequence similarity (Fuchs et al., 1999), is as yet unclear.
In this study, we investigated whether β4 interacts with the ABDs of actin-binding proteins other than plectin. We also studied the influence of the stretch of amino acids preceding the ABD on β4 binding, focusing on plectin-1A and -1C, because these two variants are strongly expressed in keratinocytes (Fuchs et al., 1999). In addition, we endeavored to identify the residues that are critically involved in the binding of the plectin-ABD to β4.
MATERIALS AND METHODS
Cell Lines and Antibodies
The immortalized keratinocyte cell lines derived from PA-JEB and MD-EBS patients have been described previously (Schaapveld et al., 1998; Geerts et al., 1999). These keratinocyte cell lines were maintained in keratinocyte serumfree medium (Life Technologies, Rockville, MD) supplemented with 50 μg/ml bovine pituitary extract, 5 ng/ml EGF, 100 U/ml penicillin, and 100 U/ml streptomycin. Rat embryo fibroblasts (REF52) and COS-7 cells were grown in DMEM (Life Technologies) containing 10% fetal calf serum (FCS).
REF52 cells and MD-EBS keratinocytes were transiently transfected with cDNA constructs using Lipofectin (Life Technologies) according to the manufacturer's instructions. COS-7 cells were transiently transfected with cDNA constructs using the DEAE-dextran method (Seed and Aruffo, 1987).
The rabbit polyclonal antibodies against the extracellular domain of β4 (H-101), the IL2-Rα (N-19) and the hemagglutinin (HA)-epitope (Y-11), and the mAb 12CA5 against the HA-epitope were purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA) The mAb 346–11A against β4 was purchased from PharMingen (San Diego, CA). The rabbit polyclonal antibody against filamin-B was a kind gift from Dr. S. Shapiro (Thomas Jefferson University, Philadelphia, PA), and the rabbit polyclonal antibody (P2) against plectin was a generous gift from Dr. H. Herrmann (German Cancer Research Center, Heidelberg, Germany). Donkey anti-rabbit horseradish peroxidase–conjugated antibody was purchased from Amersham Biosciences. (Piscataway, NJ), and FITC-conjugated goat anti-mouse antiserum from Rockland (Gilbertsville, PA). Goat anti-rabbit and anti-mouse Texas Red–conjugated antibodies and Alexa 568–conjugated phalloidin were obtained from Molecular Probes (Eugene, OR).
cDNA Constructs
All nucleotide and amino acid positions have been given a number with the ATG initiation codon at position 1. Plasmid inserts were generated by PCR, using the proofreading Pwo DNA polymerase (Roche Molecular Biochemicals, Indianapolis, IN) and gene-specific sense and antisense primers containing restriction site tags. All plasmid inserts were verified by sequencing, and protein expression and their size were confirmed by Western blotting.
The GAL4 fusion plasmids used in this study are depicted in Figures 2 and 7C. The construction of β41115–1457 and plectin-1C1–339, fused in-frame to the GAL4 activation domain (AD) of the pACT2 vector (Clontech, Palo Alto, CA), and of plectin-1C1–339, plectin-Δ165–339, dystrophin1–337, plectin-1C1–65/dystrophin11–337, and dystonin-21–336, fused in-frame to the GAL4 DNA-binding domain (BD) of the pAS2.1 vector (Clontech), has been described previously (Geerts et al., 1999; Fontao et al., 2001). Plectin-1A1–312 was generated from a human keratinocyte cDNA library by PCR using appropriately synthesized pairs of oligonucleotide primers with BamHI and SalI sites at the 5′ and 3′ end and cloned into the corresponding site of the pAS2.1 vector. In a similar way, a chimeric β4/plectin construct (unrelated sequence [URS]1–65/plectin65–339) was created by inserting a PCR-amplified cDNA fragment of β4753–818 in front of plectin-Δ165–339. Exons 1A and 1C of the PLEC1 gene were amplified by PCR, using plectin 1A1–312 and plectin 1C1–339 as templates. The various plectin point mutants were generated by the PCR overlap extension method. Dystonin-11–388 was obtained by RT-PCR on mRNA isolated from SK-N-MC cells (ATCC HTB-10), and filamin-A1–365, filamin-B1–338, utrophin1–363, and α-actinin1–337 by PCR using full-length cDNAs for these proteins as templates. The full-length cDNA for utrophin was kindly provided by Dr. S.J. Winder (University of Glasgow, Glasgow, UK) and that for α-actinin was a kind gift from Dr. D.R. Critchley (University of Leicester, Leicester, UK). Chimeras of the filamin-A and -B, utrophin, and α-actinin ABDs with the peptides encoded by exon 1A or 1C of the PLEC1 gene were generated by cloning the exonic sequences of 1A or 1C in front of the cDNAs for these ABDs. The plectin-1A1–38/dystrophin11–337 chimera was made by replacing the exonic 1C sequence in plectin-1C1–65/dystrophin11–337 with that of exonic 1A.
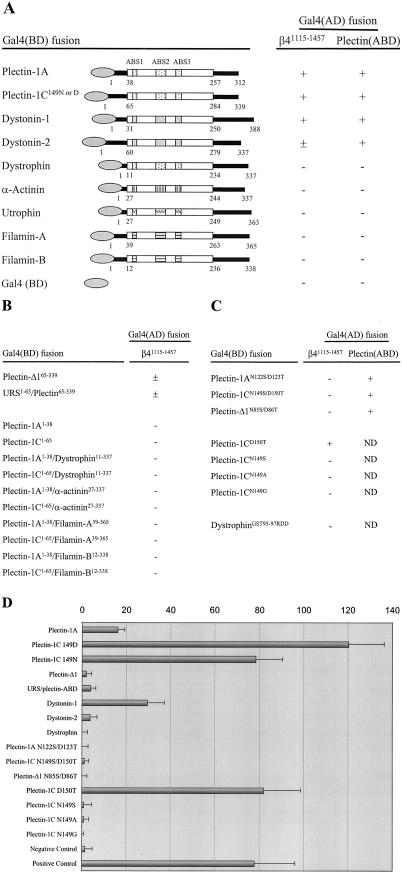
Figure 2.
Yeast two-hybrid analysis of the interactions between β4 and ABDs of several proteins. (A) Binding of ABDs of plectin, dystonin, dystrophin, α-actinin, utrophin and filamin to either β4 or the plectin-1C ABD. (B) Influence of the sequences preceding the plectin-ABD on the interaction of the ABD with β4. (C) Effect of mutations in the plectin-ABD or dystrophin-ABD on binding to β4 or the plectin-ABD. Interactions in (A–C) were scored (+), when the plating efficiencies on selective SC-LTHA plates were greater than 30% of those on nonselective SC-LT plates at 5 d of growth, (±), when they were 10–30%, and (–) when no colonies were detected at 5 d of growth or when the growth on SC-LTHA plates was <3% of that on SC-LT plates at 10 d. ND indicates not determined. (D) Quantitative β-galactosidase assay showing the strength of interactions between the various ABDs and β4 in yeast. The values indicated are arbitrary values and representatives of multiple assays. The negative control is represented by the interaction after cotransfection of β4 in pACT2 and a mock pAS2.1 vector. The positive control is represented by the interaction between PTP1–1 and PVA3–1. URS is unrelated sequence.
Figure 7.
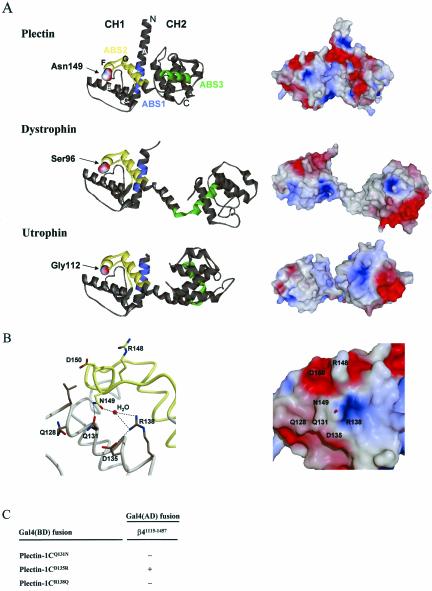
Comparison of the crystal structures and molecular surfaces of the ABDs from plectin, dystrophin and utrophin. (A) Ribbon diagrams and solvent-accessible surfaces of the ABDs of plectin, dystrophin and utrophin were created using the atomic coordinates derived by x-ray diffraction analysis (Keep et al., 1999; Norwood et al., 2000; García-Alvarez et al., 2003) and the program WebLab ViewerLite 3.20 (Molecular Simulations Inc.) for the ribbon diagrams and the program SPOCK (The Center for Macromolecular Design, Texas A&M University) for the solvent-accessible surface, colored according to electrostatic potential with values ranging from –12 kT/e- (dark red) to + 12 kT/e- (dark blue). The locations of N149 in plectin-1C and the corresponding amino acids in dystrophin and utrophin, as well as the CH1 and CH2 domains are indicated. The ABS1 is colored in blue, ABS2 in yellow, and ABS3 in green. (B) Close-up of the structure and solvent accessible surface around N149, created with SPOCK and rendered with RASTER3D (Merritt and Bacon, 1997), as in A. N149 forms the center of a shallow grove surrounded by polar residues Q128, Q131, D135, R138, R148, and D150. In the crystal structure, a water molecule occupies the small cavity. Most of the surrounding residues are conserved between plectin and dystonin, but not dystrophin, utrophin, α-actinin, and filamins A and B. The ABS2 region, colored in yellow, partially overlaps with the β4 binding pocket. (C) Effect of mutations in the plectin-ABD on binding to β4 with yeast two-hybrid analysis. Indications are as in Figure 2.
Full-length plectin-1C in pcDNA3-HA, a derivative of the eukaryotic expression vector pcDNA3 (Invitrogen Corp., Carlsbad, CA) that contains an extra sequence 5′ of the multiple cloning site encoding the HA tag, was assembled in several steps. First, the fragments 1–296 and 284–606 were generated by PCR using plectin-1C1–339 and pPLEC (a kind gift from Dr. T. Magin, University of Bonn, Germany) as templates. These products were then fused by overlap extension PCR and cloned into pcDNA3-HA, resulting in plectin-1C1–606. Subsequently, plectin-1C1–2532 was obtained by cloning an EcoRI-BstEII fragment from the pPLEC clone and a fragment of plectin2443–2532 obtained by PCR on a keratinocyte cDNA library using primers containing BstEII and BamHI restriction sites, into the EcoRI/BamHI site of plectin-1C1–606. The final fragment plectin2532–4574 was isolated from pPLEC by sequential digestion with EcoRI, Klenow fragment and HindIII. This fragment was then ligated into plectin-1C1–2532, by using XbaI (with blunt end) and HindIII restriction sites, generating a full-length plectin-1C1–4574 cDNA clone. To construct the full-length plectin-1A and plectin-Δ1 cDNA constructs, PCR fragments of plectin-1A1–312 and plectin-Δ165–339 were amplified with an EcoRV site in the upstream primers and digested with EcoRV. Subsequently, the EcoRV fragment from plectin-1C1–4574, using the EcoRV-site at position 332 of the ABD DNA, was replaced with the PCR derived EcoRV fragments of plectin-1A and -Δ1. Plectin-1C1–339 in pcDNA3-HA has been described previously (Geerts et al., 1999). The other plectin ABD constructs were prepared by exchanging EcoRV fragments as described above. Dystrophin1–337 in pcDNA3-HA was generated by PCR and subsequent cloning into pcDNA3-HA. Full-length plectin-1C/dystrophin-ABD was generated by exchanging the plectin-ABD in full-length plectin-1C with the dystrophin-ABD.
The IL2R/β4cyto chimera in pCMV has been described previously (Nievers et al., 1998).
The plectin-ABD constructs were isolated from pAS2.1 plectin-ABD (described above) and inserted into the bacterial GST-fusion protein expression vector pRP261, a derivative of the pGEX-3X vector (Amrad Corp. Ltd., Melbourne, Australia) that contains a slightly modified multiple cloning site for the production of recombinant GST fusion proteins.
Immunohistochemistry
Cryosections of mouse epithelial tissue (5–6-μm thick) placed on glass slides were fixed for 5 min in acetone at –20°C. Nonspecific binding was blocked by incubating the sections for 45 min in phosphate-buffered saline (PBS) containing 2% bovine serum albumin (BSA). After rinsing in PBS, the slides were incubated with primary antibodies, washed three times with PBS, and subsequently incubated with secondary antibodies. The sections were washed again and coverslips were then mounted onto the glass slides with VectaShield antifade (Vector Laboratories, Inc., Burlingame, CA).
RT-PCR
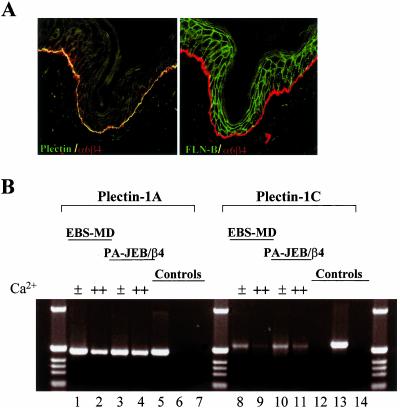
Immortalized PA-JEB/β4 (PA-JEB cells in which β4 has been reconstituted by retroviral introduction; Sterk et al., 2000) and MD-EBS keratinocytes were grown for 3 d in keratinocyte SFM, supplemented with bovine pituitary extract and EGF (low Ca2+ medium) or in a 1:3 mixture of Ham's F12 and DMEM supplemented with 5% (vol/vol) FCS (high Ca2+ medium). RNA was isolated using RNA-Bee (Tel-test, Inc., Friendswood, TX) and cDNA was made using Superscript reverse transcriptase (Invitrogen Corp.). The cDNA was used for PCR with plectin-1A– or -1C–specific primers.
Yeast Two-Hybrid Assay
Yeast strain S. cerevisiae PJ69–4A (a gift from Dr. P. James, University of Wisconsin, Madison, WI), which contains the genetic markers trp1–901, leu2–3, his3–200, gal4Δ, gal80Δ, LYS2::GAL1-HIS3, and GAL2-ADE2 (James et al., 1996), was used as the host for the two-hybrid assay. It contains two tightly regulated reporter genes, his and ade, making it suitable for the sensitive detection of protein-protein interactions. The use of PJ69–4A was essentially as described by Schaapveld et al. (1998) and Geerts et al. (1999). Equal aliquots of transformed cells were spread out on plates containing yeast synthetic complete medium lacking leu and trp (vector markers; SC-LT) or lacking leu, trp, his, and ade (vector and interaction markers; SC-LTHA). The plates were scored after 5 and 10 d of growth at 30°C. The plating efficiencies on SC-LTHA plates, compared with the plating efficiency on SC-LT plates was used as a measure of the strength of the signal generated by the two-hybrid interaction. Expression of the fusion proteins was analyzed by immunoblotting with antibodies against the GAL4 Activation Domain (AD) or GAL4 DNA-binding Domain (BD; sc-1663 and sc-510, respectively; Santa Cruz Biotechnology). As a quantitative method to measure the strength of interactions, we used a yeast β-galactosidase assay kit (Pierce Chemical, Rockford, IL), essentially as described by the manufacturer. In short, three times 5 yeast colonies were picked from the SC-LTHA plates if possible, otherwise from the SC-LT plates and grown to OD660 of 0.6–0.8 in SC-LT medium. After measuring the OD, 100 μl of the cultures was pipetted in triplo into a 96-well plate, and 100 μl of the β-galactosidase assay mixture was added. OD405 was measured at several time points.
Immunofluorescence Microscopy
MD-EBS keratinocytes, grown on glass coverslips, were transfected with HA-tagged constructs and switched to Ham's F12/DMEM (1:3) containing 5% (vol/vol) FCS, 24 h before incubation with antibodies. The cells were fixed with freshly prepared 1% (wt/vol) paraformaldehyde in PBS for 10 min at room temperature and permeabilized with 0.5% (vol/vol) Triton X-100 in PBS for 5 min at room temperature. After rinsing in PBS and blocking with 2% (wt/vol) BSA in PBS for 60 min at room temperature, the cells were incubated with primary antibodies (rabbit anti-β4 and mouse anti-HA) in PBS containing 2% BSA for 45 min at room temperature. After washing with PBS, cells were incubated with FITC-labeled anti-mouse IgG and Texas-Red–labeled anti-rabbit IgG for 45 min at room temperature.
REF52 cells were transfected with HA-tagged plectin-ABD constructs. Immunolabeling was essentially as described above for MD-EBS cells, except that the cells were fixed with 3% (wt/vol) paraformaldehyde in PBS. Instead of rabbit anti-β4 and Texas-Red–labeled anti-rabbit IgG, Alexa 568–conjugated phalloidin was used in order to stain F-actin.
After rinsing in PBS, the coverslips were mounted onto glass slides in Mowiol mounting medium (Calbiochem, San Diego, CA) containing 2.5% DABCO (Sigma-Aldrich, St. Louis, MO). Immunofluorescence images were taken using a Leica confocal laser scanning microscope.
In Vitro Binding Assay and Immunoblotting
COS-7 cells were transiently transfected with IL2R/β4cyto chimera and different plectin-HA–tagged ABD constructs. Thirty-two hours after transfection, cells were lysed with m-Per buffer (Pierce), containing a cocktail of protease inhibitors (Sigma-Aldrich, St. Louis, MO). After the lysates were cleared by centrifugation at 14,000 × g for 10 min at 4°C, mAb 12CA5 was added and the mixture was incubated o/n at 4°C. GammaBind G Sepharose (Amersham Biosciences), preincubated with BSA to block nonspecific binding sites, were incubated with the lysates for 1 h at 4°C. The beads were washed three times with m-Per buffer, dissolved in SDS-sample buffer, and analyzed by SDS-PAGE. Proteins were transferred to Immobilon-PVDF membranes (Millipore Corp., Bedford, MA) and after incubation with 2% nonfat dried milk, dissolved in TBS containing 0.05% Tween-20, to block nonspecific binding sites, the blots were incubated with first and secondary antibodies. As a substrate for the horseradish peroxidase enzyme, SuperSignal West Dura (Pierce) was used.
Purification of Recombinant Fusion Proteins
The Escherichia coli strain BL21(DE3) (Novagen, Madison, WI) was transformed with different recombinant pRP261 plasmids. Colonies obtained were used to inoculate Luria Bertani medium containing 100 μg/ml ampicillin, and cultures were grown overnight at 37°C. Cultures were then diluted 1:20 in fresh medium, grown to an OD600 of 0.7 at 37°C, and induced by the addition of isopropyl-1-thio-β-d-galactopyranoside (IPTG) to 0.2 mM overnight at 25°C. Bacteria were harvested by centrifugation at 4000 × g, resuspended in column buffer (50 mM Tris-HCl, pH 7.6, 100 mM NaCl, 1 mM EDTA, 0.1% [vol/vol] Triton X-100, 10% [vol/vol] glycerol and a cocktail of protease inhibitors), and subjected to sonication. Lysates were cleared by centrifugation for 30 min at 10,000 × g and 4°C, and the supernatants were incubated with glutathione Sepharose beads (Amersham Biosciences). Beads with affinity-bound proteins were washed with column buffer, equilibrated with elution buffer (50 mM Tris-HCl, pH 8.0, 100 mM NaCl, 1 mM EDTA, 0.1% [vol/vol] Triton X-100, 10% [vol/vol] glycerol and protease inhibitors), and eluted in elution buffer containing 10 mM glutathione.
Buffers containing the eluted fusion-proteins were exchanged in PBS containing 5% glycerol by dialysis, and proteins were concentrated using Centricon 10 filters (Millipore Corp.).
Actin-binding Assay
Before assaying the binding of recombinant proteins to F-actin, they were clarified by centrifugation at 150,000 × g for1hat24°C and kept on ice in PBS. Actin cosedimentation assays were performed with a Nonmuscle Actin Binding Protein Biochem kit (Cytoskeleton Inc., Denver, CO) as described by the supplier. In brief, actin was allowed to polymerize for 1 h at room temperature. Actin filaments were incubated with the different GST-plectin-ABD proteins for 30 min and subsequently pelleted by centrifugation at 150,000 × g for 1.5 h at 24°C. Equal amounts of pellet and supernatant were resolved by SDS-PAGE, and proteins were visualized by Coomassie Brilliant Blue staining.
RESULTS
β4 Binds to the ABDs of Plectin and Dystonin, But Not to Those of Dystrophin, Utrophin, α-actinin, and Filamin
The ABD of plectin is a domain of 220-residues with sequence similarity to the ABDs found in dystrophin, utrophin, α-actinin, and filamins. Several of these ABD containing proteins are expressed in the same cells that also express β4. Yet only plectin is colocalized with β4 in hemidesmosomes, suggesting that not all ABDs bind to β4. As shown in Figure 1A, only plectin and not, e.g., filamin-B is colocalized with β4 in basal keratinocytes. To test directly which of the ABDs of the above proteins bind to β4, we isolated their cDNAs as well as that of dystonin, which, like plectin, is a member of the plakin family and whose ABD is highly similar to that of plectin. We isolated the ABDs of splice variants of dystonin and plectin, which have different N-terminal sequences, i.e., dystonin-1 and -2 (Brown et al., 1995), and plectin-1A and -1C (Fuchs et al., 1999). Both plectin-1A and -1C are expressed in murine keratinocytes, and transcripts for both were also detected in two human keratinocyte cell lines, PA-JEB/β4 and MD-EBS, grown under low- as well as high-Ca2+ conditions (Figure 1B). The polypeptides encoded by the different cDNAs were tested in a yeast two-hybrid interaction assay against a β4 fragment containing the first pair of fibronectin type III repeats (FNIII) and the complete connecting segment (β41115–1457). In the analysis, we included a plectin-ABD construct, which contains an amino acid substitution at position 149 (N149D), because it occurs as part of a polymorphism in plectin (McLean et al., 1996; Liu et al., 1996) As shown in Figure 2A, only the ABDs of plectin and dystonin interacted with β4. Differences, however, were observed in the strength of binding, the ABD of plectin-1C149N and -1C149D binding more strongly than that of plectin-1A and dystonin-1, which in turn bound more strongly than that of dystonin-2 (Figure 2D).
Figure 1.
Localization of plectin and filamin-B in mouse skin sections and plectin transcript expression in human keratinocyte cell lines. (A) Sections were stained for β4 (red) and plectin (green, left panel) or filamin-B (green, right panel). Colocalization appears as yellow. (B) PCR with plectin-1A (lanes 1–6) and -1C specific primers (lanes 7–12) was performed on cDNA synthesized from mRNA of MD-EBS (lanes 1, 2, 8 and 9) and PA-JEB/β4 cells (lanes 3, 4, 10, and 11), grown in low (0.09 mM, ±) or high Ca2+ (1 mM, ++) as indicated. Positive and negative controls were PCRs on plasmids encoding plectin-1A (lanes 5 and 12) or plectin-1C (lanes 6 and 13). As a further negative control, no DNA was used (lanes 7 and 14).
In summary, only the ABDs of plectin and dystonin bind to β4, whereas those of dystrophin, utrophin, α-actinin, and filamin do not.
Influence of the Residues Preceding the Plectin-ABD on β4 Binding
The ABDs of plectin-1A and -1C are identical as well as those of dystonin-1 and -2. However, they bind to β4 with different affinities (Figure 2D), suggesting that the sequences preceding the ABD affect the binding activity. To further study the influence of the N-terminal sequences on the binding of the plectin-ABD to β4, the effects of removal of these sequences or their substitution by an unrelated sequence of the same length as the one encoded by exon 1C of the PLEC1 gene were analyzed. Moreover, we tested whether the peptides encoded by exons 1A and 1C could mediate β4 binding, either by themselves or when fused to the ABDs of dystrophin, α-actinin, filamin-A, or filamin-B. The data show that after the deletion or the replacement of the N-terminal sequences of plectin-1C by an unrelated sequence, binding to β4 was reduced and even weaker than that of plectin-1A (Figure 2, B and D). However, neither the fragments encoded by exons 1A and 1C, by themselves, nor when fused with the above proteins bound to β4 (Figure 2B). From these data we conclude that the stretch of amino acids that precede the ABD of plectin-1A and -1C do not bind to β4 themselves, but modulate the binding activity of the plectin-ABD, at least as assessed in yeast two-hybrid assays.
Introduction of an N149S/D150T Mutation in the Plectin-ABD Abrogates Binding to β4
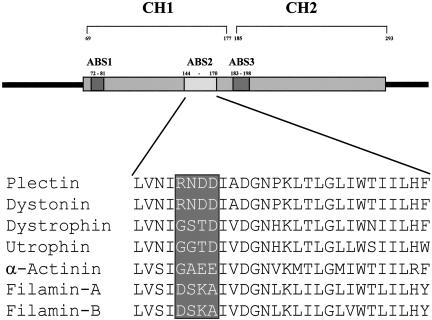
We have shown previously that binding of β4 and F-actin to plectin is mutually exclusive (Geerts et al. 1999). Because the ABS2 of the ABD was shown to be essential for binding to F-actin (Bresnick et al., 1990, 1991), we focused on this part of plectin to identify the residues that mediate binding to β4. Alignment of the ABS2 sequences from different actin-binding proteins, revealed the presence of a subregion of four amino acids that is conserved in the ABS2 of plectin and dystonin, but not in that of the other actin-binding proteins (Figure 3). A double point mutation was generated, based on the difference between the ABDs of plectin and dystrophin. The substitutions N149S/D150T (corresponding to plectin-1C numbering) were introduced in the ABDs of plectin-1A and -1C, and in the ABD construct lacking the N-terminal sequences (plectin-Δ1). In all three constructs, the double point mutation completely abolished the interaction with β4 (Figure 2, C and D). Further analysis of mutated ABDs revealed that N149, but not D150, is the critical residue for binding to β4. Substitution of N149 by serine, alanine, or glycine, as present at this position in dystrophin, utrophin, and α-actinin, reduced binding to β4 dramatically, whereas substitution of D150 by threonine, as present at this position in dystrophin, had no effect (Figure 2, C and D). Previous studies have shown that the plectin-ABD can interact with the ABD of another plectin molecule, but not with the ABD of dystrophin (Fontao et al., 2001). As shown in Figure 2, A and C, this ability of the plectin-ABD to form homodimers was not impaired by the double point mutation, indicating that residues other than those involved in β4 binding mediate binding of two plectin-ABDs to each other. Furthermore, these results show that the plectin-ABD mutant is expressed in yeast cells and thus that the lack of β4 binding was not caused by a defect in protein expression. Importantly, binding to β4 was not induced when the GST (95–97) residues in the corresponding region of the ABD of dystrophin were replaced by RDD (148–150 in plectin-1C; Figure 2C), suggesting that additional residues, other than N/D149, in the plectin-ABD are likely to be involved in binding to β4.
Figure 3.
Schematic representation of the plectin-ABD with an alignment of the amino acid sequence of the ABS2 of different ABDs. The boxed region represents a stretch of four amino acids, which is not conserved among the ABDs of plectin, dystonin, dystrophin, utrophin, α-actinin, and filamin.
Biochemical Characterization of the Plectin-ABD Variants
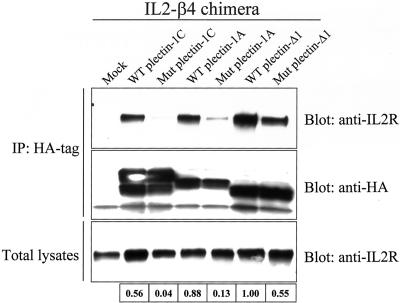
To confirm biochemically that the double point mutation in the plectin-ABD abrogates binding to β4, COS-7 cells were transfected with different HA-tagged plectin-ABD constructs (wild-type or the N149S/D150T mutant) along with an IL2R/β4cyto chimera. This IL2R/β4cyto chimera is expressed at the cell surface independent of association with α6 (Nievers et al., 1998). As shown in Figure 4, the IL2R/β4cyto chimera was coprecipitated with the ABD of both wild-type plectin-1A and -1C, as well as that of plectin-Δ1. In contrast to the results of the yeast β-galactosidase assays, we found that the ABD of plectin-Δ1 (lacking the N-terminal residues) binds most strongly to β4. Possibly, in the yeast two-hybrid assays, the bulky GAL4 binding domain, by steric interference, prevents efficient binding of the plectin-Δ1 ABD to β4. Coprecipitation of the IL2R/β4cyto chimera with the N149S/D150T mutant plectin-ABDs was strongly reduced compared with that of the wild-type ABDs (Figure 4), but was not completely abrogated. Binding of the mutated plectin-Δ1 ABD was still fairly strong, but because the binding of the ABD of wild-type plectin-Δ1 is much stronger than that of plectin-1A and -1C, the reduction of binding also in this case was considerable.
Figure 4.
Biochemical analysis of the interaction of β4 with wild-type or mutant plectin-ABDs. The top two panels show precipitation of HA tagged plectin-ABD and coprecipitation of IL2R/β4cyto. The bottom panel shows total lysates after transfection, indicating transfection efficiencies. The numbers indicate the relative strength of interaction between IL2R/β4cyto and the plectin-ABDs, with 1 representing the strongest interaction. The relative strength was calculated by determining the percentage of coprecipitated IL2R/β4cyto, compared with the total IL2R/β4cyto and correcting this value for the amount of precipitated HA-plectin-ABD. WT, wild-type; Mut, mutant (N149S/D150T)
In conclusion, these results show that the substitution of asparagine and aspartate at position 149 and 150 in the ABS2 of the plectin-ABD by serine and threonine, respectively, significantly weakens, but does not completely abrogate binding to β4.
Cell Biological Characterization of the Plectin-ABD Variants
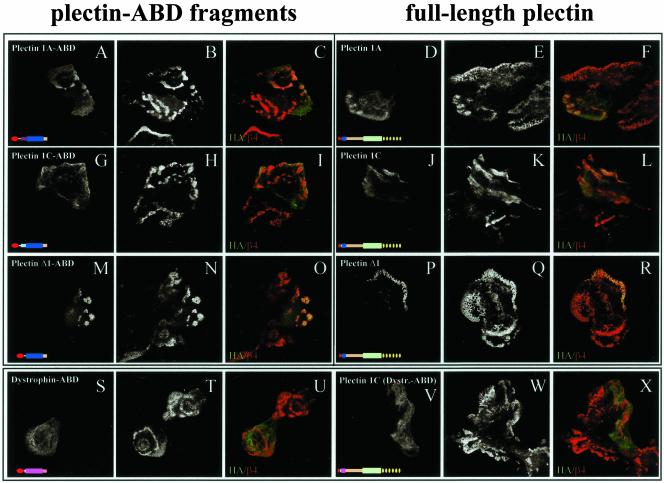
To determine the effect of the double point mutation on the colocalization of the isolated plectin-ABDs and of full-length plectin with β4 in hemidesmosomes, an HA-tagged plectin-ABD and full-length plectin (wild-type or mutant) were expressed in keratinocytes from an MD-EBS patient, who is homozygous for an 8-base pair duplication mutation in exon 31 of the PLEC1 gene, causing an early stop-codon to be introduced 14 base pairs downstream of the insertion (Smith et al., 1996). Therefore, plectin variants containing the roddomain (encoded by exon 31) are not expressed in these cells. However, we did observe the expression of a rod-less plectin variant, containing the N-terminal ABD and C-terminal sequences, which is also colocalized with β4 in hemidesmosomes (our unpublished results).
Transfection of these cells with the wild-type ABDs of plectin-1A (Figure 5, A–C), plectin-1C (Figure 5, G–I), and plectin-Δ1 (Figure 5, M–O) results in weak colocalization of these ABDs with β4 in a minority of the cells. The weak and restricted colocalization with β4 is probably due to the high affinity of the ABDs for F-actin. When full-length plectin-1A (Figure 5, D–F), plectin-1C (Figure 5, J–L), or plectin-Δ1 (Figure 5, P–R) were introduced, the colocalization with β4 was more pronounced, which is likely due to the presence of additional binding sites on plectin outside the ABD (Rezniczek et al., 1998; our unpublished results). In accordance with the results of the in vitro binding assay, we found that the recruitment of the ABD of plectin-Δ1 into hemidesmosomes is more efficient than that of the naturally occurring plectin variants, 1A and 1C. Furthermore, only the plectin-ABD and not the dystrophin-ABD was colocalized with β4 (Figure 5, S–U). A full-length chimera in which the plectin-ABD was replaced by the dystrophin-ABD, however, was weakly colocalized with β4, which also indicates the presence of additional, weak binding site(s) on plectin outside the ABD (Figure 5, V–X).
Figure 5.
Distribution of plectin-ABDs in MD-EBS keratinocytes. MD-EBS keratinocytes were transfected with HA-tagged plectin-1A ABD (A–C), full-length plectin-1A (D–F), plectin-1C ABD (G–I), full-length plectin-1C (J–L), plectin-Δ1 ABD (M–O), full-length plectin-Δ1 (P–R), dystrophin ABD (S–U) or plectin-1C1–65/dystrophin-ABD11–337/plectin339–4574 chimera (V–X). Cells were stained for HA-tagged proteins (A, D, G, J, M, P, S, V) and β4 (B, E, H, K, N, Q, T, W). Overlay images are shown in C, F, I, L, O, R, U, and X. Colocalization appears as yellow.
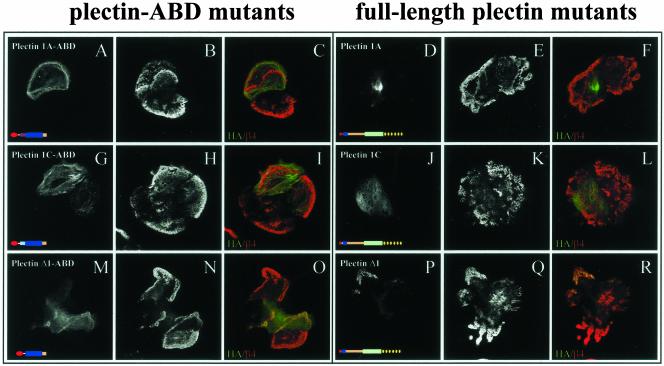
Next, the ability of plectin-1A, -1C, and -Δ1, containing the N149S/D150T substitutions in the ABS2 of the ABD, to become colocalized with β4 in MD-EBS keratinocytes was tested. In agreement with our finding that binding was strongly reduced because of these substitutions (Figure 4), colocalization of the plectin-1A, -1C, and -Δ1 N149S/D150T mutants, either as short ABD-fragments or as full-length proteins, with β4 was also reduced, but not entirely absent (Figure 6). The remaining colocalization was most evident for those mutants, whose corresponding wild-type polypeptides had the strongest basic binding activity, i.e., N149S/D150T plectin-ABD and full-length proteins, lacking the stretch of amino acids N-terminal of the ABD. Apparently, the residual binding activity of the mutant ABDs for β4 allows this weak colocalization with β4 in hemidesmosomes, or alternatively, the mutant plectin-ABDs are incorporated into hemidesmosomes by dimerization with the ABD of the endogenous rod-less plectin present in the MD-EBS cells.
Figure 6.
Distribution of mutant (N149S/D150T) plectin-ABDs in MD-EBS keratinocytes. MD-EBS keratinocytes were transfected with HA-tagged mutant plectin-1A ABD (A–C), full-length mutant plectin-1A (D–F), mutant plectin-1C ABD (G–I), full-length mutant plectin-1C (J–L), mutant plectin-Δ1 ABD (M–O), or full-length mutant plectin-Δ1 (P–R). Cells were stained as described in Figure 5.
In conclusion, the cell biological data confirm the yeast two-hybrid and biochemical findings that the plectin-ABD specifically mediates binding to β4 and that N149 is involved in this interaction.
Mapping of the Mutated Residues onto the Three-dimensional Structure of the Plectin-ABD
Mapping of the substituted amino acid of the ABS2 of the plectin-ABD (N149 in plectin-1C) onto the three-dimensional crystal structure of the plectin-ABD (García-Alvarez et al., 2003) showed that this amino acid is located at the beginning of helix F of the CH1 domain (Figure 7A). Because the fold of the CH1 domain of the ABD of plectin, (García-Alvarez et al., 2003) is almost identical to that of dystrophin (Norwood et al., 2000) and utrophin (Keep et al., 1999), it is not to be expected that substitution of N149 by either glycine or serine, as present at this position in dystrophin and utrophin, would cause a major disruption of the 3D structure (Figure 7A). Therefore, it is improbable that the loss of binding to β4 is due to a distortion of the 3D structure of the ABD. Rather, N149 may contribute directly to the binding activity by contacting one or more residues on β4. As shown in Figure 7B, N149 is structurally well defined and accessible on the surface in the center of a shallow grove surrounded by polar residues including Q128, Q131, D135, R138, R148, and D150. The side chain of N149 forms a hydrogen bond with the side chain of Q131 and together with R138 coordinates a water molecule, located in a small cavity formed by these residues and D135 (García-Alvarez et al., 2003; Figure 7B). Interestingly, like N149, Q131, D135, and R138 are conserved in dystonin (R138 being a lysine in dystonin), but not in dystrophin, utrophin, α-actinin, and the filamin isoforms. With the exception of D135, substitution of these residues (Q131 and R138) by the amino acids present at this position in dystrophin abrogates binding to β4 in a yeast two-hybrid assay (Figure 7C). This finding further supports the idea that these amino acids form a small binding pocket for a side chain of a β4 residue.
In summary, next to N149 also Q131 and R138 in the plectin-ABD are important for binding to β4, and mutation of these amino acids does not seem to affect the structure of the ABD.
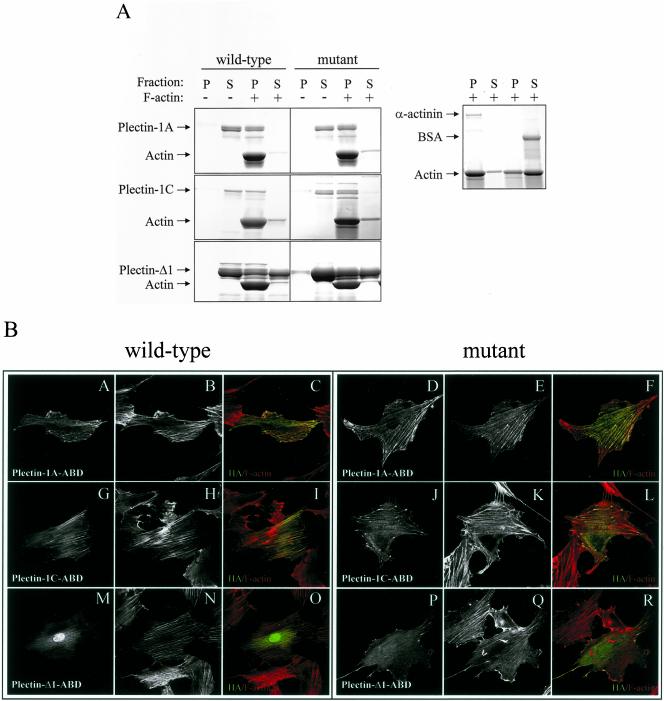
The Plectin-ABD Mutants Can Bind Actin Filaments
We have shown previously that plectin has a functional ABD, capable of binding F-actin, both in vitro and in vivo (Fontao et al., 2001). To investigate whether the double point mutation (N149S/D150T) in the ABD of plectin affects the capacity of this domain to bind F-actin, we performed an actin cosedimentation assay (Figure 8A). Consistent with the findings of Fontao et al. (2001), the results of this assay showed that the plectin-Δ1 ABD only poorly bound to F-actin, as shown by the presence of plectin-Δ1 in the supernatant (Figure 8A, bottom panel). We found that the ABDs of both plectin-1A and -1C bind F-actin with similar efficiency (Figure 8A, top and middle panels). Like the ABDs of wild-type plectin-1A, -1C and -Δ1, those carrying the N149S/D150T mutations were able to bind F-actin (Figure 8A).
Figure 8.
Biochemical analysis of the interaction of F-actin with wild-type or mutant plectin-ABDs and distribution of wild-type and mutant plectin-ABDs in REF52 cells. (A) The wild-type and mutant (N149S/D150T) plectin-ABDs were incubated with or without F-actin and centrifuged at high speed. Equal amounts of pellet (P) and supernatant (S) were subjected to SDS-PAGE and visualized by Coomassie Blue staining. (B) REF52 cells were transfected with HA-tagged wild-type (A–C, G–I, and M–O) or mutant forms (D-F, J–L, and P–R) of plectin-1A ABD (A–F), plectin-1C ABD (G–L), and plectin-Δ1 ABD (M–R). Cells were stained for HA-tagged proteins (A, D, G, J, M, P) and F-actin (B, E, H, K, N, Q). Overlay images are shown in C, F, I, L, O, and R. Colocalization appears as yellow.
The distribution of the plectin-ABDs was also assessed in transfected REF52 cells, which do not express β4. In agreement with the results of the in vitro binding assay, all constructs, wild-type and mutant, are colocalized with actin stress fibers (Figure 8B). As expected, the plectin-Δ1 wild-type and mutant constructs became colocalized with F-actin less efficiently than the plectin-1A and -1C constructs (Figure 8B).
These results show that the mutated ABDs, whose capacity to bind β4 is strongly reduced, bind to F-actin as efficiently as the wild-type ABDs, further implicating that the fold of the ABD is not affected by the mutation.
DISCUSSION
In this study we show that the ABD of plectin, and also that of dystonin, interacts with the integrin β4 subunit and that the residues preceding the ABD dramatically influence the affinity of binding, although they themselves do not bind to β4. Furthermore, we have identified three residues that are critical for tight binding of the plectin-ABD to β4. One of these amino acids, N149 (as numbered in plectin-1C) is located in the ABS2 of the plectin-ABD, whereas the other two (Q131 and R135) reside in the region preceding this sequence. These amino acids are not present in the ABDs of dystrophin, utrophin, and filamin-A and -B, and the ABDs of these proteins do not interact with β4.
Characteristics of the ABD for Binding to β4 Integrin
Plectin contains an ABD of the β-spectrin type, which is highly conserved among other ABD-containing proteins, such as dystonin, dystrophin, α-actinin, utrophin, and filamins. Several of these ABD-containing proteins are expressed in basal keratinocytes that also express α6β4. However, of these proteins, only plectin is recruited by β4 into hemidesmosomes in keratinocytes. Our results, which show that β4 binds to the plectin-ABD, but not to the ABDs of dystrophin, α-actinin, and filamin-A and -B, explain why plectin is the only of these proteins that is present in hemidesmosomes. Interestingly, also the dystonin-ABD interacts with β4. Other than plectin, dystonin is not expressed in keratinocytes, but in neuronal cells (Brown et al., 1995). Initially, dystonin was cloned and characterized as the neuronal variant of BP230, encoded by the BPAG1 gene. It was reported that dystonin (or BPAG1-n), like BP230 (or BPAG1-e), contains a plakin domain, a central rod-domain and C-terminal plakin repeats containing an intermediate filament-binding domain as well as an N-terminal ABD (Brown et al., 1995). However, recent studies indicated that not BPAG1-n, but BPAG1-a is the predominant product of the BPAG1 gene in neuronal cells. BPAG1-a shares the ABD and plakin domains with BPAG1-n, but its rod domain is composed of spectrin repeats and the C-terminal domain contains a Gas2-related microtubule-binding domain (Leung et al., 2001). Possibly, in previous studies on dystonin, BPAG1-a and not BPAG1-n was investigated. We only studied the N-terminal part of dystonin, which is present in both BPAG1-n and BPAG1-a. Dystonin was reported to be essential for the polarization, matrix attachment, and organization of the cytoskeleton in Schwann cells during myelination (Bernier et al., 1998). Interestingly, α6β4 also is expressed in Schwann cells (Sonnenberg et al., 1990; Einheber et al., 1993; Niessen et al., 1994), its expression requiring continuous axon-Schwann cell interactions and its localization is only polarizing during myelination (Feltri et al., 1994). Feltri and coworkers already suggested a possible function of α6β4 in providing polarity and communication of the basal lamina with the cytoskeleton in order to generate the mechanical force necessary for Schwann cell myelination. However, the possibility of physical interactions between α6β4 and intermediate filaments (vimentin, nestin, or neurofilaments) and/or microtubules in Schwann cells had to be explored. Our current findings suggest that in Schwann cells it may be dystonin/BPAG1-a that links the α6β4 integrin to the cytoskeleton.
The Effect of the Residues Preceding the Plectin-ABD
The PLEC1 gene encodes a wide variety of plectin splice variants. Only some of these variants are expressed in keratinocytes, most strongly plectin-1A and -1C and more weakly, plectin-1 and -1B (Fuchs et al., 1999). Recently, Andrä et al. (2003) reported that only plectin-1A is present in basal keratinocytes of the mouse and recruited into hemidesmosomes, whereas plectin-1C is not. However, the PA-JEB and MD-EBS keratinocyte cell lines, which are derived from human basal keratinocytes, contain in addition to plectin-1A also plectin-1C transcripts, albeit at lower levels (Figure 1B). Because we lack splice variant-specific antibodies, we cannot test whether both variants are expressed at the protein level or whether they both are located in hemidesmosomes. Our results with the ABD of plectin, in which the N-terminal sequence of residues had been deleted or exchanged with an unrelated sequence, confirm that the residues preceding the ABD can affect its affinity for β4. Furthermore, in the yeast two-hybrid assay plectin-1C bound to β4 with much higher efficiency. However, although in both the in vitro binding assay and the immunofluorescence experiment the binding of plectin-1A to β4 appeared to be slightly more efficient than binding of plectin-1C to β4, the difference in β4 binding between plectin-1A and -1C in these experiments was not significant. Most likely, other factors in the cell play an important role in the binding of plectin to β4, such as the binding of plectin to other hemidesmosomal components or a regulation of its affinity for F-actin and β4 by posttranslational modifications. Furthermore, in both MD-EBS and PA-JEB/β4 cells dimerization of the exogenously introduced ABDs with the ABD of endogenous plectin may play a role in binding to β4 and thus their recruitment into hemidesmosomes (Fontao et al., 2001).
The N-terminal sequences of amino acids do not only influence the binding of the plectin-ABD with β4, but also the binding of plectin to F-actin. Deletion of the N-terminal residues specific for plectin-1A or -1C results in decreased binding of the plectin-ABD to F-actin, as evaluated in an actin-polymerization assay and in transfection experiments with REF52 cells. Similar observations were made with the ABD of utrophin, whose affinity for F-actin is reduced by a factor four after deletion of the N-terminal residues (Keep et al., 1999). Interestingly, although deletion of the N-terminal sequences reduces the affinity of the plectin-ABD to F-actin, it increases the binding activity for β4. This is suggested by the observation that plectin constructs lacking the sequences N-terminal of the ABD localized more efficiently into hemidesmosomes than the corresponding plectin-1A and -1C constructs. Thus, it seems that binding of the plectin ABD to either F-actin or β4 can be regulated via the N-terminal sequence of amino acids in an apparently opposite manner, i.e., a decrease in binding to F-actin and an increase in binding to β4, and vice versa. Further work is needed to identify the signals that regulate this affinity switch.
The Effect of Mutating N149 in Plectin on β4 Binding
Our observations that a double point mutation in the ABS2 of the plectin-ABD is sufficient to completely abrogate binding to β4 in yeast, but not to prevent its colocalization with β4 in transfected MD-EBS cells, cannot be explained by the possibility that we have introduced a putative phosphorylation site. In yeast, phosphorylation of serine at this site may result in a different binding affinity of plectin for β4. However, by substituting the critical asparagine residue at position 149 by alanine or glycine instead of serine, we exclude such a mechanism. Importantly, a complete loss of binding of the plectin-ABD mutants to β4, as observed in the yeast two-hybrid assay, was not found in the in vitro binding assay. Together these findings indicate that the substituted amino acid at position 149, although important for binding to β4, is not solely responsible for the interaction of the plectin-ABD with β4. Indeed, we found that also glutamine and arginine at positions 131 and 138, respectively, are critical for binding to β4. Furthermore, an asparagine residue at position 149 of the plectin-ABD permits binding to β4, but also an aspartate residue allows β4 binding. Together these results indicate that multiple residues on the plectin-ABD are involved in the binding to β4.
Structure of the Plectin-ABD
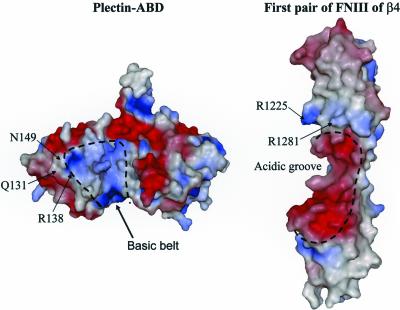
García-Alvarez et al. (2003) observed a basic “belt” in plectin around the interdomain waist and the helix G of CH1, which is surrounded by acidic residues, one of which is N149 (Figure 9). The shape of the basic belt is such that it fits well in the acidic V-shaped groove formed at the interface between the first and second FNIII repeat of β4 (de Pereda et al., 1999). Interestingly, the residues R1225 and R1281 of β4 that are critical for binding to the plectin-ABD (Koster et al., 2001) lie at the edge of this acidic groove (Figure 9). García-Alvarez et al. (2003) already suggested that the plectin-β4 interaction involves the interdomain surfaces in both pairs of tandem CH and FNIII domains. If so, either R1225 or R1281 in β4 may come into close proximity with Q131, R138, or N149 in plectin-1C, allowing a direct contact between one or more of these amino acids.
Figure 9.
Solvent-accessible surfaces of the plectin-ABD and the first pair of FNIII repeats of β4 colored as described in Figure 7. The basic cleft in the plectin-ABD extends along the interdomain contact, and is flanked by negatively charged patches. The acidic groove in β4 has shape complementarity to the basic belt in the plectin-ABD and is flanked by positively charged residues.
In the crystal structure of the plectin-ABD, the residues Q131, R138, and N149, together with D135 form a small cavity in which a well-ordered water molecule is present. There is direct contact between N149 and Q131, and between R138 and D135, whereas R138 and N149 are involved in coordinating the water molecule located in this cavity (García-Alvarez et al., 2003; Figure 7B). Possibly, this water molecule is displaced when a side chain from a β4 residue docks into the pocket. Alternatively, the water molecule may form part of the complex and mediate a specific contact with β4. Interestingly, only the residues forming contacts with either N149 (Q131) or the water molecule (R138), but not D135, which contacts R138, are crucial for binding to β4. Mutation of residue D150 that like the above residues, is available at the surface of the structure, but located somewhat outside the putative binding pocket, does not affect binding to β4 in a yeast two-hybrid assay (Figure 2C). Additionally, other regions in the ABD may be involved in the interaction with β4. The interaction requires both CH domains of plectin (Geerts et al., 1999), suggesting that CH2 harbors additional contact sites. The first pair of FNIII domains of β4 adopts an extended conformation in the crystal structure (de Pereda et al., 1999), and no significant interdomain bending is likely to occur upon binding to plectin, because the linker between the FNIII domains is very short. Therefore the binding interface in plectin is likely to extend over only one of the faces of the ABD. The area of CH2 around the ABS3 is oriented on the same side of the molecule as Q131, R138, and N149 (Figure 7A). In fact the ABS2-ABS3 face of the ABD contains most of the solvent exposed residues conserved among plectin and dystonin but not in the other ABDs. Thus, the ABS2-ABS3 face of the plectin-ABD, including N149, is presumably the side of the plectin-ABD involved in binding to β4. The fact that N149 is part of the β4 binding pocket but also of the ABS2, and the close proximity of this pocket to ABS2, explains how by steric interference the binding of the plectin-ABD to β4 and F-actin is mutually exclusive.
Comparison of the crystal structures of the ABDs of plectin (García-Alvarez et al., 2003), utrophin (Keep et al., 1999), and dystrophin (Norwood et al., 2000; Figure 7A), shows that the fold of the CH1 domain is very similar in these ABDs. Hence, the substitution of Q131, R138, or N149 for residues as they occur in utrophin or dystrophin, most likely does not induce large structural differences. However, the position of the CH2 domain relative to the CH1 domain is different in the crystal structures of these three ABDs (Figure 7A) because of the dimerization by domain swapping of the ABDs of utrophin and dystrophin. Despite the high degree of structural conservation in the backbone of the CH1 and CH2 domains, the solvent exposed residues are not conserved among the three ABDs, leading to specific surface details in each molecule. As a consequence, the electrostatic surfaces of the molecules are different (Figure 7A). Therefore, lack of binding to β4 by ABDs other than plectin and dystonin may be due to loss of specific contacts, loss of charge complementarity, or variations in the relative orientation of CH domains. The presence of multiple interaction sites within the ABD may explain why the substitution of the three amino acids GST (position 95–97) in the ABS2 of dystrophin by RDD as present in plectin, does not induce binding to β4. The structure of the amino acid stretch preceding the ABD is not modeled. Because we found the stretch of amino acids preceding the ABD to be important in regulating the binding of the plectin-ABD to β4, we hypothesize that these residues may spatially interfere with the binding site for β4, e.g., the basic belt or the surrounding acidic residues.
Acknowledgments
We thank Dr. P. James for the yeast strain PJ69–4A, Dr. T Magin for pPLEC, Dr. S.J. Winder for the utrophin cDNA, and Dr. D.R. Critchley for the human α-actinin cDNA. We thank Dr. S. Shapiro and Dr. H. Herrmann for the generous gifts of antibodies. We gratefully acknowledge Dr. A. Perrakis, Dr. E. Danen, and Prof. C.P.E. Engelfriet for critically reading the manuscript. This work was supported by grants from the Dutch Cancer Society (NKI 99-2039) and the Dystrophic Epidermolysis Bullosa Research Association (DEBRA Foundation, Crowthorne, UK).
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E03–05–0268. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E03-05-0268.
Abbreviations used: ABD, actin binding domain; ABS, actin binding sequence; BP, bullous pemphigoid; CH, calponin homology domain, FNIII, fibronectin type III repeat; MD-EBS, muscular dystrophy associated with epidermolysis bullosa simplex; PA-JEB; pyloric atresia associated with junctional epidermolysis bullosa.
References
- Andrä, K., Kornacker, I., Jorgl, A., Zorer, M., Spazierer, D., Fuchs, P., Fischer, I., and Wiche, G. (2003). Plectin-isoform-specific rescue of hemidesmosomal defects in plectin (–/–) keratinocytes. J. Invest. Dermatol. 120, 189–197. [DOI] [PubMed] [Google Scholar]
- Andrä, K., Lassmann, H., Bittner, R., Shorny, S., Fässler, R., Propst, F., and Wiche, G. (1997). Targeted inactivation of plectin reveals essential function in maintaining the integrity of skin, muscle, and heart cytoarchitecture. Genes Dev. 11, 3143–3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernier, G., De Repentigny, Y., Mathieu, M., David, S., and Kothary, R. (1998). Dystonin is an essential component of the Schwann cell cytoskeleton at the time of myelination. Development 125, 2135–2148. [DOI] [PubMed] [Google Scholar]
- Borradori, L., and Sonnenberg, A. (1996). Hemidesmosomes: roles in adhesion, signaling and human diseases. Curr. Opin. Cell Biol. 8, 647–656. [DOI] [PubMed] [Google Scholar]
- Bresnick, A.R., Janmey, P.A., and Condeelis, J. (1991). Evidence that a 27-residue sequence is the actin-binding site of ABP-120. J. Biol. Chem. 266, 12989–12993. [PubMed] [Google Scholar]
- Bresnick, A.R., Warren, V., and Condeelis, J. (1990). Identification of a short sequence essential for actin binding by Dictyostelium ABP-120. J. Biol. Chem., 265, 9236–9240. [PubMed] [Google Scholar]
- Brown, A., Dalpe, G., Mathieu, M., and Kothary, R. (1995). Cloning and characterization of the neural isoforms of human dystonin. Genomics 29, 777–780. [DOI] [PubMed] [Google Scholar]
- Burgeson, R.E., and Christiano, A.M. (1997). The dermal-epidermal junction. Curr. Opin. Cell Biol. 9, 651–658. [DOI] [PubMed] [Google Scholar]
- de Pereda, J.M., Wiche, G., and Liddington, R.C. (1999). Crystal structure of a tandem pair of fibronectin type III domains from the cytoplasmic tail of integrin α6β4. EMBO J. 18, 4087–4095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowling, J., Yu, Q.C., and Fuchs, E. (1996). β4 integrin is required for hemidesmosome formation, cell adhesion and cell survival. J. Cell Biol. 134, 559–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Einheber, S., Milner, T.A., Giancotti, F., and Salzer, J.L. (1993). Axonal regulation of Schwann cell integrin expression suggests a role for α6β4 in myelination. J. Cell Biol. 123, 1223–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feltri, M.L., Scherer, S.S., Nemni, R., Kamholz, J., Vogelbacker, H., Scott, M.O., Canal, N., Quaranta, V., and Wrabetz, L. (1994). β4 integrin expression in myelinating Schwann cells is polarized, developmentally regulated and axonally dependent. Development 120, 1287–1301. [DOI] [PubMed] [Google Scholar]
- Foisner, R., and Wiche, G. (1987). Structure and hydrodynamic properties of plectin molecules. J. Mol. Biol. 198, 515–531. [DOI] [PubMed] [Google Scholar]
- Fontao, L., Geerts, D., Kuikman, I., Koster, J., Kramer, D., and Sonnenberg, A. (2001). The interaction of plectin with actin: evidence for cross-linking of actin filaments by dimerization of the actin-binding domain of plectin. J. Cell Sci. 114, 2065–2076. [DOI] [PubMed] [Google Scholar]
- Fuchs, P., Zorer, M., Rezniczek, G.A., Spazierer, D., Oehler, S., Castanon, M.J., Hauptmann, R., and Wiche, G. (1999). Unusual 5′ transcript complexity of plectin isoforms: novel tissue-specific exons modulate actin binding activity. Hum. Mol. Genet. 8, 2461–2472. [DOI] [PubMed] [Google Scholar]
- Gache, Y., Chavanas, S., Lacour, J.P., Wiche, G., Owaribe, K., Meneguzzi, G., and Ortonne, J.P. (1996). Defective expression of plectin/HD1 in epidermolysis bullosa simplex with muscular dystrophy. J. Clin. Invest. 97, 2289–2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Alvarez, B., Bobkov, A., Sonnenberg, A., and de Pereda, J.M. (2003). Structural and functional analysis of the actin binding domain of plectin suggests alternative mechanisms for binding to F-actin and integrin α6β4. Structure 11, 615–625. [DOI] [PubMed] [Google Scholar]
- Geerts, D., Fontao, L., Nievers, M.G., Schaapveld, R.Q., Purkis, P.E., Wheeler, G.N., Lane, E.B., Leigh, I.M., and Sonnenberg, A. (1999). Binding of integrin α6β4 to plectin prevents plectin association with F-actin but does not interfere with intermediate filament binding. J. Cell Biol. 147, 417–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georges-Labouesse, E., Messaddeq, N., Yehia, G., Cadalbert, L., Dierich, A., and Le Meur, M. (1996). Absence of integrin α6 leads to epidermolysis bullosa and neonatal death in mice. Nat. Genet. 13, 370–373. [DOI] [PubMed] [Google Scholar]
- Gimona, M., Djinovic-Carugo, K., Kranewitter, W.J., and Winder, S.J. (2002). Functional plasticity of CH domains. FEBS Lett. 513, 98–106. [DOI] [PubMed] [Google Scholar]
- Giudice, G.J., Emery, D.J., and Diaz, L.A. (1992). Cloning and primary structural analysis of the bullous pemphigoid autoantigen BP180. J. Invest. Dermatol. 99, 243–250. [DOI] [PubMed] [Google Scholar]
- Green, K.J., and Jones, J.C. (1996). Desmosomes and hemidesmosomes: structure and function of molecular components. FASEB J. 10, 871–881. [DOI] [PubMed] [Google Scholar]
- Hartwig, J.H. (1994). Actin-binding proteins 1, spectrin superfamily. Protein Profile 1, 706–778. [PubMed] [Google Scholar]
- James, P., Halladay, J., and Craig, E.A. (1996). Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144, 1425–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, J.C., Kurpakus, M.A., Cooper, H.M., and Quaranta, V. (1991). A function for the integrin α6β4 in the hemidesmosome. Cell Regul. 2, 427–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keep, N.H., Winder, S.J., Moores, C.A., Walke, S., Norwood, F.L., and Kendrick-Jones, J. (1999). Crystal structure of the actin-binding region of utrophin reveals a head-to-tail dimer. Structure Fold. Des. 7, 1539–1546. [DOI] [PubMed] [Google Scholar]
- Koster, J., Kuikman, I., Kreft, M., and Sonnenberg, A. (2001). Two different mutations in the cytoplasmic domain of the integrin β4 subunit in nonlethal forms of epidermolysis bullosa prevent interaction of β4 with plectin. J. Invest. Dermatol. 117, 1405–1411. [DOI] [PubMed] [Google Scholar]
- Leung, C.L., Zheng, M., Prater, S.M., and Liem, R.K. (2001). The BPAG1 locus: Alternative splicing produces multiple isoforms with distinct cytoskeletal linker domains, including predominant isoforms in neurons and muscles. J. Cell Biol. 154, 691–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine, B.A., Moir, A.J., Patchell, V.B., and Perry, S.V. (1992). Binding sites involved in the interaction of actin with the N-terminal region of dystrophin. FEBS Lett. 298, 44–48. [DOI] [PubMed] [Google Scholar]
- Liu, C.G., Maercker, C., Castanon, M.J., Hauptmann, R., and Wiche, G. (1996). Human plectin: organization of the gene, sequence analysis, and chromosome localization (8q24). Proc. Natl. Acad. Sci. USA 93, 4278–4283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean, W.H. et al. (1996). Loss of plectin causes epidermolysis bullosa with muscular dystrophy: cDNA cloning and genomic organization. Genes Dev. 10, 1724–1735. [DOI] [PubMed] [Google Scholar]
- Merritt, E.A., and Bacon, D.J. (1997). Raster 3D: Photorealistic molecular graphics. In: Methods in Enzymology, Macomolecular Cryptallography, Part B. ed. C.W. Carter and R.M. Sweet, New York: Academic Press, 277, 505–524. [DOI] [PubMed] [Google Scholar]
- Nakano, A. et al. (2001). Epidermolysis bullosa with congenital pyloric atresia: novel mutations in the β4 integrin gene (ITGB4) and genotype/phenotype correlations. Pediatr. Res. 49, 618–626. [DOI] [PubMed] [Google Scholar]
- Niessen, C.M., Cremona, O., Daams, H., Ferraresi, S., Sonnenberg, A., and Marchisio, P.C. (1994). Expression of the integrin α6β4 in peripheral nerves: localization in Schwann and perineural cells and different variants of the β4 subunit. J. Cell Sci. 107, 543–552. [DOI] [PubMed] [Google Scholar]
- Nievers, M.G., Schaapveld, R.Q., Oomen, L.C., Fontao, L., Geerts, D., and Sonnenberg, A. (1998). Ligand-independent role of the β4 integrin subunit in the formation of hemidesmosomes. J. Cell Sci. 111, 1659–1672. [DOI] [PubMed] [Google Scholar]
- Norwood, F.L., Sutherland-Smith, A.J., Keep, N.H., and Kendrick-Jones, J. (2000). The structure of the N-terminal actin-binding domain of human dystrophin and how mutations in this domain may cause Duchenne or Becker muscular dystrophy. Structure Fold. Des. 8, 481–491. [DOI] [PubMed] [Google Scholar]
- Pulkkinen, L., Rouan, F., Bruckner-Tuderman, L., Wallerstein, R., Garzon, M., Brown, T., Smith, L., Carter, W., and Uitto, J. (1998). Novel ITGB4 mutations in lethal and nonlethal variants of epidermolysis bullosa with pyloric atresia: missense versus nonsense. Am. J. Hum. Genet. 63, 1376–1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rezniczek, G.A., de Pereda, J.M., Reipert, S., and Wiche, G. (1998). Linking integrin α6β4-based cell adhesion to the intermediate filament cytoskeleton: direct interaction between the β4 subunit and plectin at multiple molecular sites. J. Cell Biol. 141, 209–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruzzi, L., Gagnoux-Palacios, L., Pinola, M., Belli, S., Meneguzzi, G., D'Alessio, M., and Zambruno, G. (1997). A homozygous mutation in the integrin α6 gene in junctional epidermolysis bullosa with pyloric atresia. J. Clin. Invest. 99, 2826–2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaapveld, R.Q. et al. (1998). Hemidesmosome formation is initiated by the β4 integrin subunit, requires complex formation of 4 and H.D1/plectin, and involves a direct interaction between β4 and the bullous pemphigoid antigen 180. J. Cell Biol. 142, 271–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seed, B., and Aruffo, A. (1987). Molecular cloning of the CD2 antigen, the T-cell erythrocyte receptor, by a rapid immunoselection procedure. Proc. Natl. Acad. Sci. USA 84, 3365–3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, F.J. et al. (1996). Plectin deficiency results in muscular dystrophy with epidermolysis bullosa. Nat. Genet. 13, 450–457. [DOI] [PubMed] [Google Scholar]
- Sonnenberg, A. et al. (1991). Integrin α6β4 complex is located in hemidesmosomes, suggesting a major role in epidermal cell-basement membrane adhesion. J. Cell Biol. 113, 907–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnenberg, A., Linders, C.J., Daams, J.H., and Kennel, S.J. (1990). The α6β1 (VLA-6) and α6β4 protein complexes: tissue distribution and biochemical properties. J. Cell Sci. 96, 207–217. [DOI] [PubMed] [Google Scholar]
- Steinbock, F.A., and Wiche, G. (1999). Plectin: a cytolinker by design. Biol. Chem. 380, 151–158. [DOI] [PubMed] [Google Scholar]
- Stepp, M.A., Spurr-Michaud, S., Tisdale, A., Elwell, J., and Gipson, I.K. (1990). α6β4 integrin heterodimer is a component of hemidesmosomes. Proc. Natl. Acad. Sci. USA 87, 8970–8974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterk, L.M., Geuijen, C.A., Oomen, L.C., Calafat, J., Janssen, H., and Sonnenberg, A. (2000). The tetraspan molecule CD151, a novel constituent of hemidesmosomes, associates with the integrin α6β4 and may regulate the spatial organization of hemidesmosomes. J. Cell Biol. 149, 969–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Neut, R., Krimpenfort, P., Calafat, J., Niessen, C.M., and Sonnenberg, A. (1996). Epithelial detachment due to absence of hemidesmosomes in integrin β4 null mice. Nat. Genet. 13, 366–369. [DOI] [PubMed] [Google Scholar]
- Vidal, F., Aberdam, D., Miquel, C., Christiano, A.M., Pulkkinen, L., Uitto, J., Ortonne, J.P., and Meneguzzi, G. (1995). Integrin β4 mutations associated with junctional epidermolysis bullosa with pyloric atresia. Nat. Genet. 10, 229–234. [DOI] [PubMed] [Google Scholar]
- Wiche, G. (1998). Role of plectin in cytoskeleton organization and dynamics. J. Cell Sci. 111, 2477–2486. [DOI] [PubMed] [Google Scholar]