Abstract
The Net2, Fis1, and Dnm1 proteins are required for the division of mitochondria in the yeast Saccharomyces cerevisiae. Net2p has an amino-terminal region that contains predicted coiled-coil motifs and a carboxyl-terminal domain composed of WD-40 repeats. We found that the amino-terminal part of Net2p interacts with Fis1p, whereas the carboxyl-terminal region interacts with both Dnm1p and Fis1p. Overproduction of either domain of Net2p in yeast cells poisons mitochondrial fission, and the dominant-negative effect caused by the WD-repeats of Net2p is suppressed by increased levels of Dnm1p. Point mutations in the WD-region of Net2p or in the GTPase region of Dnm1p disrupt the normal Net2p-Dnm1p interaction, causing Net2p to lose its normal punctate distribution. Our results suggest that Dnm1p interacts with the WD-repeats of Net2p and in a GTP-dependent manner recruits Net2p to sites of mitochondrial division. Furthermore, our results indicate that Net2p is required for proper assembly of the mitochondrial fission components to regulate organelle division.
INTRODUCTION
Mitochondria function in a number of diverse cellular processes including ATP synthesis, lipid metabolism, and programmed cell death (Tzagoloff, 1983; Attardi and Schatz, 1988; Gottlieb, 2000). In many eukaryotic cells, mitochondria are elongated, branched organelles that are distributed throughout the cell. The shape and number of mitochondria appears to be regulated in response to a variety of environmental cues and developmental programs (Bereiter-Hahn and Voth, 1994; Hales and Fuller, 1997; Hermann and Shaw, 1998; Labrousse et al., 1999; Jensen et al., 2000). In vegetatively growing Saccharomyces cerevisiae cells, the number and shape of mitochondria seems to be controlled by a balance of fusion and fission events. In addition, regulated division and fusion appear to play key roles in mitochondrial dynamics during meiosis and sporulation (Miyakawa et al., 1984).
Mitochondrial division in yeast is mediated by at least three proteins: Dnm1p, Net2p (also called Gag3p; Fekkes et al., 2000), Mdv1p (Tieu and Nunnari, 2000), and Fis2p (Mozdy et al., 2000), and Fis1p (also called Mdv2p; Tieu and Nunnari, 2000). Cells lacking any of these three proteins contain a single mitochondrion consisting of a network of interconnected tubules. The Dnm1 protein was originally identified based on its homology to dynamin, a protein required for endocytosis (Gammie et al., 1995). Much of Dnm1p is found in small dot-like structures on the mitochondrial surface, often observed at sites of division (Bleazard et al., 1999; Sesaki and Jensen, 1999). However, an appreciable fraction of a Dnm1p-GFP fusion protein can be detected in small, fast moving structures in the cytosol, but the role of this portion of Dnm1p remains unclear (Jensen et al., 2000). During subcellular fractionation, nearly all of the Dnm1p is found in the cytosol, suggesting that the interaction between Dnm1p and the mitochondrial surface is weak and may be dynamic (Cerveny et al., 2001). Like most of the dynamin family members, Dnm1p contains an amino-terminal GTPase domain, and a recent study has shown that Dnm1p can hydrolyze GTP in vitro (Fukushima et al., 2001).
Net2p was identified as a Dnm1p-interacting protein in a genome-wide two-hybrid screen (Uetz et al., 2000). Several other studies showed that net2 mutants (called gag3, mdv1, and fis2) are defective in mitochondrial division (Fekkes et al., 2000; Mozdy et al., 2000; Tieu and Nunnari, 2000). For simplicity, we will hereafter refer to Gag3/Mdv1p/Fis2p/Net2p as Net2p. Net2p contains predicted coiled-coil regions in its amino-terminus and six to seven WD-40 repeats near its carboxyl-terminus (Tieu and Nunnari, 2000; Cerveny et al., 2001). Net2p is distributed in a punctate pattern on the mitochondrial surface (Tieu and Nunnari, 2000; Cerveny et al., 2001), with many of the Net2-containing structures colocalizing with Dnm1p at sites of mitochondrial constriction (Cerveny et al., 2001). The distribution of Net2p depends on Dnm1p, because Net2p becomes evenly dispersed along the mitochondrial outer membrane in dnm1 null cells (Tieu and Nunnari, 2000; Cerveny et al., 2001).
Fis1p is an 18-kDa protein anchored in the mitochondrial outer membrane by a carboxyl-terminal hydrophobic tail, and unlike Dnm1p and Net2p, the Fis1 protein is distributed uniformly along the mitochondrial surface (Mozdy et al., 2000). Recent studies suggest that Fis1p functions in recruiting both Dnm1p and Net2p to the mitochondrial surface, and it has been suggested that Fis1p acts early in the division event (Mozdy et al., 2000; Tieu et al., 2002). In this study we have further examined the roles that Net2p, Dnm1p, and Fis1p play in mitochondrial fission, and our results suggest that Net2p organizes fission proteins, including Dnm1p and Fis1p, into a complex that is competent for division. Additionally, our data suggest that Dnm1p plays an early and important role in recruiting Net2p to sites mitochondrial division.
MATERIALS AND METHODS
Strains, Media, and Genetic Methods
Wild-type strains FY833 and FY834 (Winston et al., 1995), MATa GAL1::lacZ strain Y190 (Bai and Elledge, 1996), MATα GAL1::lacZ strain N106r (Uetz et al., 2000), and dnm1Δ strain YHS83 (Sesaki and Jensen, 2001) have been described. net2::HIS3 strain RJ1367 was constructed by PCR-mediated gene disruption (Adams et al., 1997) in strain FY834 using oligonucleotides 642 and 643 (oligonucleotide sequences available upon request). Similarly, fis1::HIS3 strain RJ1365 was constructed using oligonucleotides 644 and 645. fis1::HIS3 net2::kanMX4 strain RJ1567 was formed by PCR disruption of NET2 in strain RJ1365. fis1::HIS3 DNM1::kanMX4 strain RJ1566 was produced by PCR disruption of DNM1 in strain RJ1365 using oligonucleotides 295 and 296.
To express the GAL1 controlled proteins, cells were grown overnight in medium with 2% raffinose (Sraf) to log phase, galactose was added to 1–2%, and cells were incubated for 180 min unless otherwise mentioned. Standard yeast media and genetic techniques were used (Adams et al., 1997).
Plasmid Construction
pOAD and pOBD were as described (Uetz et al., 2000). The construction of pOAD-DNM1 and pOBD-NET2 (Cerveny et al., 2001) and pHS20 and pHS83 (Sesaki and Jensen, 1999) was previously described.
pHS78, a plasmid that encodes a matrix-targeted cox4-RFP fusion protein was constructed as follows. The red fluorescent protein (RFP) was amplified from pDsRed.T1-N1 (gift from Ben Glick, University of Chicago) using oligonucleotides 461 and 465 and digested with XbaI and NotI. RFP was used to replace GFP in pHS12, a CEN LEU2 vector that contains ADH1-COX4-GFP (Sesaki and Jensen, 1999).
To construct pKC61, a CEN-TRP1 vector that carries a GAL1 promoted green fluorescent protein (GFP), the GFP coding sequence was PCR amplified from pAA1 (Sesaki and Jensen, 1999) using oligonucleotides 317 and 318, digested with XhoI and BamHI, and then ligated into XhoI/BamHI-cut pRS314GU (Sikorski and Hieter, 1989).
pKC62, a CEN-TRP1 plasmid that encodes a galactose-inducible GFP-Net2 fusion protein, was constructed by inserting a 2140-base pair BamHI fragment that contained NET2 from pKC5 (Cerveny et al., 2001) into BamHI-cut pKC61.
To construct pKC69, a CEN-TRP1 plasmid that carries a galactose inducible Net2p, we ligated a BamHI fragment from pKC5 (Cerveny et al., 2001) into BamHI-cut pRS314GU (Sikorski and Hieter, 1989).
To construct pKC64, a CEN-TRP1 plasmid that expresses a galactose inducible fusion between GFP and the first 386 amino acids of Net2p (Net2Np), we first amplified a 1200-base pair DNA from yeast genomic DNA using oligonucleotides 442 and 759. The PCR product was digested with BamHI and ligated into BamHI-cut pJE6 (Cerveny et al., 2001) forming pKC63. pKC63 is a CEN-LEU2 plasmid that carries a triple HA epitope fused to Net2CCp. DNA encoding Net2Np was liberated from pKC63 with BamHI and ligated into BamHI-cut pKC61 to form pKC64.
pKC65, a CEN URA3 vector that encodes a GAL1-controlled GFP-Net2Np fusion, was constructed by inserting a PvuI fragment from pKC64 into PvuI-digested pRS316 (Sikorski and Heiter, 1989).
To construct pKC67, a CEN-TRP1 plasmid that expresses a galactose-inducible fusion of GFP to the last 329 amino acids of the Net2 protein (Net2WDp), a 1025-base pair fragment was amplified from yeast genomic DNA using oligonucleotides 677 and 443. The PCR product was digested with BamHI and ligated into BamHI-cut pJE6 (Cerveny et al., 2001), forming pKC66, a CEN-LEU2 plasmid that expresses a triple HA-Net2WDp fusion. A BamHI fragment from pKC66 was ligated into a BamHI-digested pKC61 creating pKC67.
pKC68, a CEN-LEU2 plasmid that encodes a GAL1 controlled GFP-Net2WDp fusion, was constructed by ligating a PvuI fragment from pKC67 plasmid into PvuI-cut pRS315 (Sikorski and Hieter, 1989).
To construct pKC71, a CEN-TRP1 vector that expresses a galactose-inducible Net2WDp, a 1025-base pair fragment was PCR amplified from yeast genomic DNA using oligonucleotides 737 and 443, digested with BamHI, and then ligated into BamHI-cut pRS314GU (Sikorski and Hieter, 1989).
Point mutations in the WD-40 repeats of Net2p were generated using PCR-mediated site-directed mutagenesis (Quickchange, Stratagene, La Jolla, CA) of pKC62, a CEN-TRP1 plasmid that carries a galactose inducible GFP-Net2p fusion. pKC80, which encodes GFP-Net2pF400A was formed using oligonucleotides 932 and 933. pKC81, a vector encoding GFP-Net2pR461A was created with oligonucleotides 934 and 935. pKC82, a plasmid that carries a GFP-Net2pD664A fusion was generated using oligonucleotides 936 and 937. pKC83, a vector that carries a GFP-Net2pS689A fusion, was formed using oligonucleotides 938 and 939. All changes were confirmed by DNA sequencing.
pKC85, a CEN LEU2 plasmid that encodes a galactose-inducible GFP-Net2WDR461A fusion protein, was constructed by amplifying NET2WDR461A from pKC81 with oligonucleotides 677 and 443. The PCR product was digested with BamHI and inserted into BamHI-cut pKC68.
pKC53 and pKC54 are plasmids that carry S42N and T62F mutations in the GTPase region of Dnm1p, respectively. They were generated by PCR-mediated site-directed mutagenesis (Quickchange, Stratagene). Specifically, pKC51, a bacterial expression plasmid that encodes a fusion between the maltose-binding protein (MBP) and Dnm1p, was mutagenized. pKC51 was constructed by amplifying DNM1 from yeast genomic DNA with oligonucleotides 636 and 637. The resulting PCR product was digested with BamHI and SalI and then ligated into BamHI/SalI-cut pMALcRI (New England Biolabs, Beverly, MA) to create pKC51. To create pKC53, which encodes a fusion between MBP and Dnm1pS42N, codon 42 was changed from TCC (ser) to AAC (asn) using oligonucleotides 747 and 748. Likewise, pKC54, which encodes an MBP-Dnm1pT62F fusion, was formed when codon 43 was changed from ACA (thr) to TTT (phe) using oligonucleotides 749 and 750. All changes were confirmed by DNA sequencing.
CEN-LEU2 plasmids that encode DNM1S42N or DNM1T62F fused to GFP were named pKC75 and pKC76, respectively. First, oligonucleotides 268 and 790 were used to PCR amplify DNA 500 base pairs upstream of the DNM1 start site using yeast genomic DNA as template. DNM1 mutant ORFs were amplified from pKC53 or pKC54 using oligonucleotides 791 and 269. PCR products were digested and then ligated into XhoI/NotI-digested pAA1 (Sesaki and Jensen, 1999).
CEN-LEU2 plasmids that encode the DNM1 dominant mutants fused to the triple HA epitope were constructed by ligating XhoI/NotI-liberated fragments from pKC75 and pKC76 into XhoI/NotI-digested pAA3 to generate pKC78 (Dnm1pS42N-HA) and pKC79 (Dnm1pT62F-HA).
pOBD-Net2Np, a CEN-TRP1 plasmid that carries the Gal4p DNA-binding domain (GBD) fused to the first 386 amino acids of Net2p, was constructed by homologous recombination. We amplified the first 1158 base pairs of NET2 from pKC5 (Cerveny et al., 2001), using oligonucleotides 455 and 494. PvuII/NcoI-cut pOBD and PCR product were cotransformed into Y190 cells to allow homologous recombination between the gapped vector and the PCR product.
pOBD-Net2WDp, a CEN-TRP1 plasmid that carries the GBD fused to the last 328 amino acids of Net2p, was constructed by cotransforming EcoRI-cut pOBD and the final 987 base pairs of NET2, which were amplified from yeast genomic DNA using oligonucleotides 493 and 456 into yeast strain Y190 to allow homologous recombination.
pOBD-Fis1p, a CEN-TRP1 vector that carries a fusion between GBD and Fis1p was constructed by ligating the FIS1 gene and 300 base pairs of down-stream sequence that was amplified from yeast genomic DNA using oligonucleotides 701 and 792 into EcoRI/NcoI-cut pOBD.
pOAD-Fis1p, a CEN-LEU2 plasmid which encodes a fusion between the activating domain of Gal4p (GAD) and the Fis1 protein, was constructed by ligating an EcoRI-cut FIS1 PCR product that was amplified from yeast genomic DNA using oligonucleotides 701 and 702 into EcoRI-digested pOAD.
pOBD-WDpF400A, pOBD-WDpR461A, pOBD-WDpD664A, and pOBD-WDpS689A are CEN-TRP1 plasmids that encode fusions between GBD and Net2WDp point mutants. PvuII/NcoI-cut pOBD and PCR products generated using oligonucleotides 456 and 493 and pKC80, pKC81, pKC82, and pKC83, respectively, were cotransformed into N106r cells where homologous recombination occurred.
pOAD-S42N and pOAD-T62F are CEN-LEU2 plasmids that encode fusions between GAD and GTPase inactive forms of Dnm1p. Using oligonucleotides 453 and 454, we PCR amplified DNM1S42N and DNM1T62F from pKC53, and pKC54, respectively. PCR products were cotransformed with EcoRI-cut pOAD into Y190 cells where homologous recombination formed the plasmids.
To create pKC50, NET2 was amplified from yeast genomic DNA using oligonucleotides 436 and 437, digested with EcoRI and XbaI, and then ligated into EcoRI/XbaI-cut pMALcRI (New England Biolabs).
Yeast Two-Hybrid Analyses
Strain Y190 was cotransformed with a pOAD plasmid, containing a Net2p, Dnm1p, or Fis1p fused to the Gal4p activation domain and a pOBD plasmid, carrying a Net2p, Dnm1p, or Fis1p fused to the Gal4p DNA-binding region. Alternatively, Y190 cells with a pOBD construct were mated to N106r cells with a pOAD construct. β-galactosidase activity of the GAL1::lacZ reporter in Y190 and N106r cells was measured as described (Adams et al., 1997). Non-specific interactions were checked using p53 and rat lamin (BD Biosciences, Clontech, Palo Alto, CA).
Biochemical Analyses
Proteins were extracted from yeast cell as described (Yaffe and Schatz, 1984). Yeast cells were fractionated as described (Daum et al., 1982; Davis et al., 1998), except that homogenization and wash buffers contained 1 mM phenylmethylsulfonyl fluoride, 1 μg/ml pepstatin, 1 μg/ml aprotinin, 1 μg/ml leupeptin, and 1 μM trans-epoxy-succinyl-l-leucylamido (4-guanidino) butane (all from Sigma, St. Louis, MO). For strains that carry galactose-inducible proteins, cells were grown in Sraf medium supplemented with the appropriate amino acids to OD600 of 1.6–2.0. Galactose (1% final conc.) was added to the cultures and cells were grown for another 2 h.
Proteins were separated by SDS-PAGE (Laemmli, 1970), transferred to Immobilon membranes (Millipore, Bedford, MA) and Western blotted using standard methods (Haid and Suissa, 1983). For protein blotting, membranes were decorated with antiserum to GFP (A. Aiken Hobbs, unpublished data; 1:5000 dilution), Tim23p (Emtage and Jensen, 1993; 1:10.000 dilution), hexokinase (A. Davis, unpublished data; 1:20,000 dilution), Dnm1p (1:5000 dilution), or Net2p (1:10,000 dilution). Immune complexes were detected with horseradish peroxidase–conjugated secondary antibodies (Amersham Pharmacia, Piscataway, NJ) at a 1:10,000 dilution followed by enhanced chemiluminescence (SuperSignal; Pierce, Rockford, IL). Western blots were quantitated using the VersaDoc imaging system (Bio-Rad, Hercules, CA) with Quantity One software (Bio-Rad).
Coimmune Precipitations from Whole Cell Extracts
Wild-type cells (FY833) carrying pGAL1-DNM1-HA (pHS15) in combination with either pGAL1-GFP-Net2Np (pKC65) or pGAL1-GFP-Net2WDp (pKC68) were grown to early log phase in raffinose-containing medium and then shifted to galactose-containing medium for 4–6 h. Alternatively, dnm1Δ cells (YHS83) carrying pGAL1-DNM1-HA (pHS15) with either pGAL1-GFP-Net2WDp or pGAL1-GFP-Net2WDpR461A (pKC85) were grown to early log phase in raffinose, and then galactose was added to a final concentration of 2% to induce expression from the GAL1 promoter for 4 h. Cells were harvested by centrifugation, resuspended in IP buffer (0.5% Triton X-100, 150 mM NaCl, 1 mM EDTA, and 50 mM Tris-HCl, pH 7.5, protease inhibitor cocktail diluted 1:200 [P8340, Sigma]) to 20 OD600 units per ml, and lysed by vortexing with glass beads five times for 30 s. Cleared lysate, 400–500 μl, was incubated with 10 μl of HA antibodies (12CA5; Nieman et al., 1983) at 4°C with gentle rotation for 5 h. A 50% slurry of protein A-Sepharose (100 μl; Sigma) in IP buffer was added to each reaction, the samples were incubated for 1 h more, and the beads were collected by centrifugation. The supernatants were treated with 100 μl of 6× SDS-sample buffer containing β-ME and then boiled for 5 min. The protein A-Sepharose was washed, 4 times with 1 ml of IP buffer, and the bound proteins were eluted by two sequential extractions with 200 μl 6× SDS sample buffer at 65°C for 10 min. The pellet volume was adjusted to that of the supernatant with 150 μl of IP buffer containing 2% Triton X-100. Equal volumes of pellet and supernatant were analyzed by SDS-PAGE followed by Western blotting with GFP antibodies.
Antibody Generation
MBP fusions to Net2p (pKC50) or Dnm1p (pKC51) were induced in E. coli strain ER2366 (New England Biolabs) at 37°C with 40 μM IPTG for 4 h. MBP-Net2p was isolated from inclusion bodies (Harlow and Lane, 1988) and MBP-Dnm1p was purified using amylose affinity chromatography (New England Biolabs). Proteins excised from SDS-polyacrylamide gels were injected into rabbits (Covance Research, Inc., Denver, PA).
Microscopy
Yeast cells were observed during log phase growth using an Axioskop microscope with a 100× Plan-Neofluar objective (Carl Zeiss Inc., Thornwood, NY). Fluorescence and differential interference contrast (DIC) images were captured with an Orca ER camera (Hamamatsu, Bridgewater, NJ) using OpenLab software v3.0.8 (Improvision, Lexington, MA). To visualize mitochondria, cells were either treated with 10 nM mitofluor red 589 (Molecular Probes, Eugene, OR) or by expressing in cells a matrix-targeted cox4-RFP fusion protein from pHS78. Size and number of Dnm1p-GFP dots were counted and measured using OpenLab software. 3-D microscopy of cells was obtained by collecting a series (20–25) of images every 0.2 μm in the Z-axis using a Delta Vision system (Applied Precision, Issaquah, WA), which was based on an IX 70 microscope with a 100× Plan-Apo 1.32NA objective (Olympus, Melville, NY) and a CH350 CCD camera (Roper Industries, Princeton, NJ). Each image was deconvolved then projected using the Delta Vision software.
RESULTS
Net2p Contains at Least Two Functional Domains That Interact with Dnm1p and Fis1p
The Net2 protein is predicted to contain coiled-coil regions in its amino-terminus and six or seven WD40-repeats in its carboxyl-terminus (Figure 1A). To test whether these two domains carry out specific functions in the cell, we first examined their interactions with Dnm1p and Fis1p using the yeast two-hybrid system. Plasmids carrying fusions between the Gal4 DNA-binding domain and either the full-length Net2 protein (pOBD-NET2), the first 386 amino acids of Net2p (pOBD-N), or the last 328 amino acids of Net2p (pOBD-WD) were constructed. These plasmids were then tested for interactions with Dnm1p or Fis1p using vectors pOAD-DNM1 or pOAD-FIS1, encoding Dnm1p or Fis1p fused to the Gal4p activating domain. Consistent with earlier studies (Uetz et al., 2000; Tieu and Nunnari, 2000; Cerveny et al., 2001), we found that full-length Net2p interacted with Dnm1p. Significant levels of β-galactosidase activity were detected from a GAL1::lacZ reporter when cells contained both the pOBD-NET2 and pOAD-DNM1 constructs (Figure 1B).
Figure 1.
Net2p contains two functional domains. (A) The Net2 protein contains predicted coiled-coil regions (amino acids 129–150 and 230–300, hatched boxes) and WD-40 repeats (amino acids 386–714, speckled box). The Net2Np and Net2WDp domains that were used in this study are denoted. (B) Dnm1p interacts with the carboxyl-terminal WD-40 repeats of Net2p in the yeast two-hybrid assay. Relative β-galactosidase activities of a GAL1::lacZ reporter in yeast cells carrying pOAD-Dnm1p in combination with pOBD-Net2p, pOBD-WDp, or pOBD-Net2Np are shown. As controls for self-activation, cells expressing only pOBD-Net2p, pOBD-WDp, or pOBD-Np were also tested. The averages of three independent experiments are shown with error bars of one SD. (C) Fis1p interacts with both the amino-terminal and carboxyl-terminal domains of Net2p. β-galactosidase activities of cells with pOAD-Fis1p and pOBD-Net2p, pOBD-WDp, or pOBD-Net2Np are shown as in B. (D) Dnm1p and the WD-repeats of Net2p interact in vivo. WT cells (FY833) carrying pHS15 (pGAL1-DNM1-HA) and either pKC65 (pGAL1-GFP-NET2N) or pKC68 (pGAL1-GFP-WD) were lysed, and total proteins were immune-precipitated with HA antibodies. One hundred percent of the supernatant (S) and pellet (P) were analyzed by SDS-PAGE followed by Western blotting with Dnm1p or G3DPH for controls and GFP antisera to detect GFP-Net2Np or GFP-Net2WDp. Sixty percent of total protein input (I) from Net2Np-containing extracts and 32% of input from Net2WDp-containing extracts were used.
We found that the Dnm1p-interacting part of Net2p was localized to the C-terminal, WD-repeat–containing domain of Net2p (Net2WDp). β-galactosidase activity was observed in cells carrying pOAD-DNM1 and expressing Net2WDp from the pOBD-WD plasmid. No activity was detected in cells carrying pOAD-DNM1 and expressing the amino-terminal half of Net2p, Net2Np, from the pOBD-N plasmid. Our two-hybrid analyses also indicated that both parts of Net2p interact with Fis1p. Cells carrying pOAD-FIS1 and either pOBD-N or pOBD-WD produced β-galactosidase (Figure 1C). These results differ from a previous report by Tieu et al. (2002), in which the carboxyl-terminal region of Net2p was found to interact with Dnm1p, but the aminoterminal domain only interacted with Fis1p. The reasons for the dissimilar results are not clear, but we note that both our Fis1p-containing construct and our assay conditions differ from the other study. Our construct carries the carboxyl-terminal, membrane-spanning domain of Fis1p and we assayed β-galactosidase activity from a GAL1::lacZ reporter, whereas the Fis1p construct used by Tieu et al. (2002) lacked the transmembrane segment and GAL2::ADE2 activity was monitored. Also, the Net2p carboxyl-terminal constructs differed slightly. Regardless, we note that the interaction of Fis1p with Net2WDp that we detect appears to be stronger than that with Net2Np. We also found no evidence that Fis1p and Dnm1p associate directly, because no β-galactosidase activity was detected when cells contained both pOBD-FIS1 and pOAD-DNM1 (K. Cerveny, unpublished observations).
To confirm the results of our two-hybrid studies, we examined the in vivo interaction between Dnm1p and different domains of Net2p (Figure 1D). Wild-type cells carrying Dnm1p-HA in combination with GAL1 controlled versions of GFP-Net2Np or GFP-Net2WDp were collected after 4 h of galactose induction, and proteins were immune-precipitated from lysed cells with HA antibodies. Dnm1p was efficiently immune precipitated by our procedure, with more than 90% of the Dnm1p-HA found in the pellet fraction, whereas nearly all of the cytosolic protein, glyceraldehyde-3-phosphate dehydrogenase (G3DPH) was found in the supernatant (Figure 1D). In extracts from cells expressing both Dnm1p-HA and GFP-Net2Np, we found all of the GFP-tagged protein was found in the supernatant after immune precipitation of Dnm1p-HA, suggesting that Dnm1p does not tightly bind to the amino-terminal portion of Net2p. However, in extracts from cells containing both Dnm1p-HA and GFP-Net2WDp, we found that a significant amount of the carboxyl-terminal portion of Net2p coimmune precipitated with Dnm1p-HA. Approximately 30% of the Net2WDp was found in the pellet fraction along with Dnm1p-HA. These results support our yeast two-hybrid studies, indicating that Dnm1p interacts with the WD-repeats of Net2p. However, because only a fraction of the Net2WD protein precipitated with Dnm1p, we speculate that the interaction between Dnm1p and Net2WDp may be labile and/or dynamic.
Overproduction of Net2p, Net2Np, and Net2WDp Inhibit Mitochondrial Division
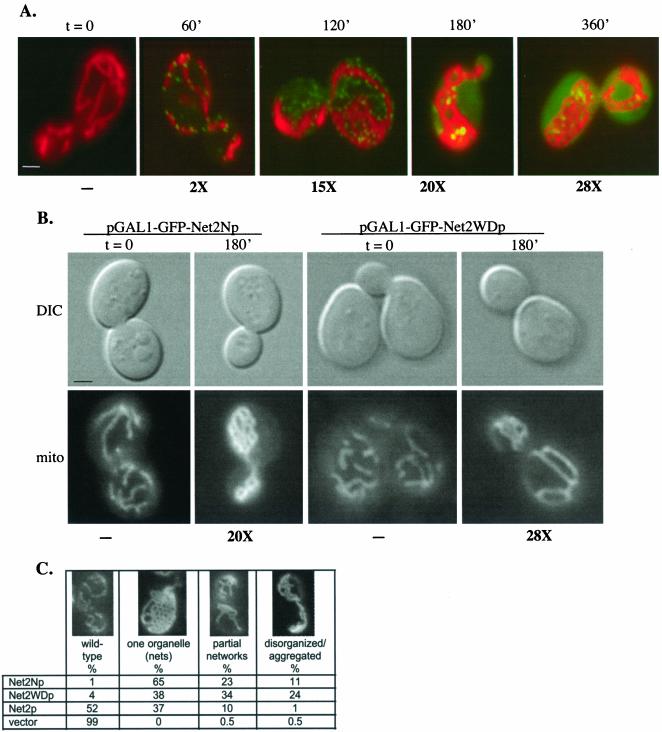
To further examine the function of the two domains of Net2p, we expressed Net2Np or Net2WDp from a galactose-inducible promoter in wild-type yeast cells. To follow the distribution of the different Net2 proteins, each was fused to the GFP. Mitochondria were detected by expressing a matrix-targeted red fluorescent protein (cox4-RFP) or by staining cells with a mitochondrial-specific fluorescent dye (mitofluor 589). As a control, we first examined the effect of expressing full-length Net2p fused to GFP. Cells carrying pGAL1-GFP-NET2 were pregrown in raffinose-containing medium and then shifted to galactose medium to induce the expression of GFP-Net2p. In addition to examining cells by fluorescence microscopy, proteins were isolated from cells at each time point, and the amount of Net2 protein was determined by quantitative Western blotting.
Until 60 min after induction, GFP-Net2p was undetectable in cells. At ∼60 min, GFP-Net2p first appeared in small, dot-like structures on the surface of the mitochondria (Figure 2A). This punctate distribution was similar to that seen in cells expressing wild-type Net2p (Cerveny et al., 2001), and the levels of Net2p approximate those of wild type. As the amounts of GFP-Net2p increased, we noticed that the number of GFP-Net2p dots first increased (Figure 2A, 120′) such that many of these dots did not appear on mitochondria. Later, much of the GFP-Net2p began to distribute uniformly throughout the cytosol, with only several larger GFP-Net2p spots visible on the mitochondria (Figure 2A, 180′ and 360′). The appearance of GFP-Net2p in the cytosol seemed to coincide with a defect in mitochondrial fission. One hundred eighty minutes after induction, ∼37% of the cells contained a single mitochondrion consisting of a network of interconnected tubules, and 12% of the cells contained fewer mitochondria, some of which were disorganized (Figure 2C). In about half of the cells (∼51%), mitochondrial number and shape appeared normal.
Figure 2.
Overproduction of Net2p, Net2Np, or Net2WDp inhibits mitochondrial division. (A) Overproduction of Net2p. Wild-type strain FY833 containing pKC62, a vector that carries galactose-inducible GFP-Net2p, were pregrown on raffinose-containing medium and then shifted to galactose for the indicated times. Cells were viewed using fluorescence microscopy after staining with mitofluor red 589, and representative cells showing merged images from the red (mitochondria) and green (GFP-Net2p) channels are shown. Beneath each frame is the level of Net2p calculated from Western blotting with Net2p antibody. Bar, 3 μm. (B) Overproduction of Net2Np or Net2WDp. WT cells containing either pKC64 (pGAL1-GFP-Net2Np) or pKC67 (pGAL1-GFP-Net2WDp) were pregrown in raffinose medium (t = 0) and then shifted to galactose (t = 180′). Cells also carry plasmid, pHS78, which produces the matrix-targeted cox4-dsRed.T1 protein. Images of cells (DIC) and the dsRed-containing mitochondria (mito) are shown. Beneath each frame is the level of Net2p determined by Western blotting. Bar, 3 μm. (C) Quantitation of mitochondrial morphology of wild-type cells overexpressing GFP-Net2p, GFP-Net2Np, or GFP-Net2WDp. WT cells containing pKC62, pKC64, or pKC67, or the empty vector, pRS314GU, were observed after 180 min of growth in galactose medium. The mitochondrial morphology of more than 100 cells of each strain was determined.
We also found that galactose-induced expression of either Net2Np or Net2WDp also inhibited mitochondrial fission in a dominant manner. Overproduction of Net2Np was the most potent inhibitor of division, with almost all of the cells containing altered mitochondria. After 180 min of induction, nearly 90% of cells carrying pGAL1-GFP-NET2N contained either a single, net-like mitochondrion (65% of cells) or a partial network of interconnected tubules (23% of cells; Figure 2, B and C). Net2WDp produced from pGAL1-GFP-NET2WD also interfered with mitochondrial division in more than 90% of the cells, but the effect was less pronounced than that of Net2Np. Only ∼38% of cells contained a single mitochondrial network, with ∼58% of cells displaying either partial networks or disorganized mitochondria (Figure 2, B and C). Western blotting showed that Net2p, Net2Np, and Net2WDp were similarly overproduced from the GAL1 promoter, with levels of each about ∼20-fold greater than wild-type Net2p after 180 min of induction (Figure 2, A and B; see supplemental data for representative Western blots).
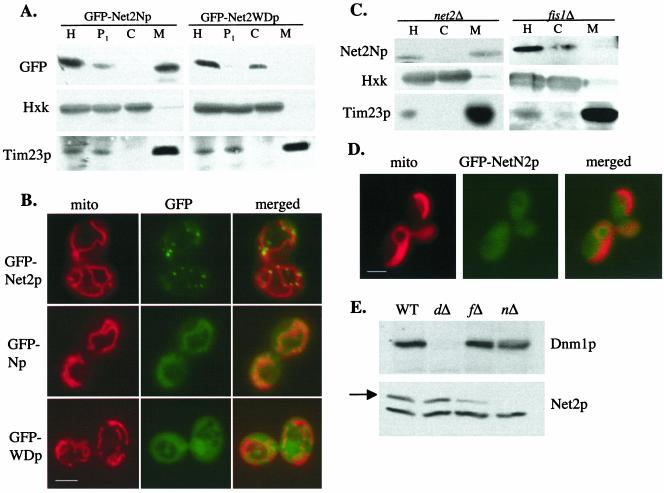
Like the wild-type Net2 protein (Tieu and Nunnari, 2000; Cerveny et al., 2001), we found that Net2Np is located on mitochondria (Figure 3A). When cells expressing GFP-Net2Np were subjected to subcellular fractionation, Western blotting showed that GFP-Net2Np was located in the mitochondrial pellet along with Tim23p, a mitochondrial inner membrane protein. In contrast to full-length Net2p, which is found in punctate structures on mitochondria, microscopy showed that GFP-Net2Np was uniformly distributed along mitochondrial tubules (Figure 3B). Consistent with previous results (Tieu et al., 2002), we found that Fis1p is important for the localization Net2Np. GFP-Net2Np fusion protein was found in the cytosol of fis1 cells (Figure 3, C and D). Interestingly, we have found that Fis1p is also needed for the stability of Net2p, because much less Net2p is present in fis1Δ cells than in wild-type cells (Figure 3E). Therefore, both the mislocalization and instability of Net2p in fis1Δ cells may contribute to the block in mitochondrial division seen in fis1Δ cells. However, expression of Net2p from the GAL1 promoter in fis1Δ cells does not appear to stimulate mitochondrial fission, suggesting that both Fis1p and Net2p may function together to mediate division of mitochondria.
Figure 3.
Localization of Net2Np and Net2WDp. (A) Net2Np is mitochondrial; Net2WDp is cytosolic. WT cells of strain FY833 containing pKC64 (pGAL1-GFP-Net2Np) or pKC67 (pGAL1-GFP-Net2WDp) were grown in galactose medium for 120 min. Cells were homogenized (H), and separated by centrifugation into a low-speed pellet (P1), a mitochondrial fraction (M), and crude cytosol (C). After SDS-PAGE, fractions were analyzed by Western blotting using antibodies to GFP (GFP-Net2Np or GFP-Net2WDp), hexokinase (Hex), and Tim23p. (B) Net2Np colocalizes with mitochondria; Net2WDp is diffusely distributed throughout the cytosol. net2Δ cells expressing GFP-Net2p from plasmid pKC62, or WT cells expressing GFP-Net2Np or GFP-Net2WDp from plasmids pKC64 or pKC67 were examined by fluorescence microscopy. Mitochondria were visualized by staining with mitofluor red 589. Bar, 3 μm. (C) Net2Np requires Fis1p for its mitochondrial localization. Cells were fractionated as in part a and homogenate (H), high-speed supernatant, crude cytosol (C), and mitochondria (M) were analyzed. (D) Location of GFP-Net2Np expressed from pKC64 for 90 min was examined in fis1Δ cells using fluorescence microscopy. Bar, 3 μm. (E) Net2p requires Fis1p for its stability. Whole cell extracts from WT, dnm1Δ (dΔ), fis1Δ (fΔ), and net2Δ (nΔ) were analyzed by SDS-PAGE and Western blotting with antibodies to Dnm1p (top) and Net2p (arrow, bottom).
Net2WDp showed a very different distribution from either Net2p or Net2Np. By both subcellular fractionation and microscopy, we found that Net2WDp was located throughout the cytosol (Figure 3, A and B), indicating that the amino terminal region of Net2p is required to target the full-length protein to mitochondria. After very long periods of induction (t [mtequ] 8 h), we noticed that GFP-Net2WDp could form a few dot-like structures near the cell periphery in addition to the diffuse cytoplasmic staining (K. Cerveny, unpublished results).
Overproduction of Dnm1p Suppresses the Dominant-negative Effect of Net2WDp
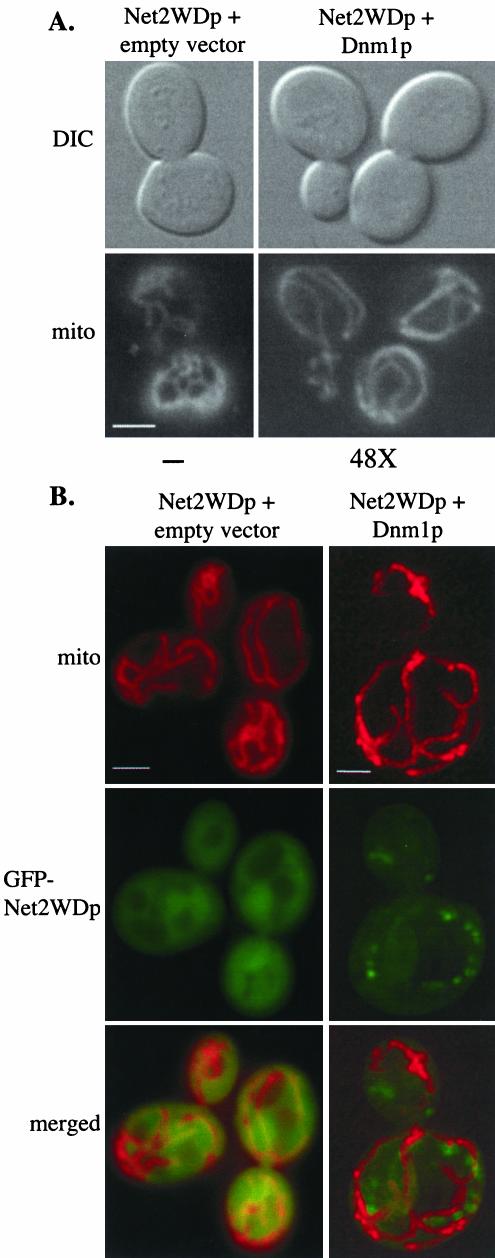
Our two-hybrid studies and coimmune precipitation experiments suggest that Dnm1p interacts directly with the C-terminal, WD-containing domain of Net2p. Further supporting this possibility, we found that the inhibition of mitochondrial fission by Net2WDp was rescued when cells also contained high levels of Dnm1p. Cells containing pGAL1-GFP-NET2WD were transformed with a plasmid carrying a galactose-inducible version of the DNM1 gene. After 240 min of induction of both Net2WDp and Dnm1p, we found that the shape and number of mitochondria were more like wild-type than in cells expressing only Net2WDp (Figure 4A). Many (68%, n = 100) of the cells with both Dnm1p and Net2WDp contained several tubular-shaped mitochondria, similar to those seen in wild-type cells. Under these conditions we found that Dnm1p presence was ∼50 times greater than that of wild-type (Figure 4A). The suppression of the dominant-negative effect of Net2WDp was limited to Dnm1p; we found that coexpression of galactose-inducible Net2p or Fis1p constructs did not significantly rescue the mitochondria in cells with Net2WDp. Furthermore, when high-copy levels of both Net2p and Fis1p were present in Net2WDp-expressing cells, a defect in mitochondrial division was still observed (K. Cerveny, unpublished observations).
Figure 4.
Overproduction of Dnm1p suppresses the dominant-negative effect of Net2WDp. (A) Cells expressing Net2WDp in combination with high levels of Dnm1p contain nearly wild-type mitochondria. Strain FY833 containing pKC68 (pGAL1-GFP-Net2WDp) was transformed with pRS314GU (empty vector) or pHS15 (pGAL1-Dnm1p-HA). Cells were viewed by DIC and fluorescence microscopy after growth on galactose medium for 180 min. Mitochondria were visualized by staining with mitofluor red 589. Beneath each frame, the level of Dnm1p overexpression is shown. Bar, 5 μm. (B) Localization and distribution of Net2WDp changes when Dnm1p is overexpressed. Wild-type (FY833) cells expressing GFP-Net2WDp from pKC67 were examined by fluorescence microscopy (Net2WDp + empty vector). Wild-type cells expressing both Dnm1p and GFP-Net2WDp from the GAL1 promoter were stained with mitofluor red 589 and examined using the Delta Vision system (Net2WDp + Dnm1p). The image shown is deconvolved, projected data that was rotated 180° around the y-axis. Bar, 5 μm.
The inhibition of mitochondrial division caused by expression of the Net2N protein was not rescued by overproduction of Dnm1p, Fis1p, or Net2p. Induction studies using strains that carried pGAL1-NET2N in combination with pGAL1-DNM1, pGAL1-FIS1, or pGAL1-NET2 showed mitochondrial morphologies similar to those with pGAL1-NET2N alone (K. Cerveny, unpublished observations). In addition, the localization and distribution of Dnm1p-GFP does not change in cells when Net2Np is expressed (K. Cerveny, unpublished results). Thus, the dominant-negative effect of Net2Np may arise by its interaction with some as yet unknown protein required for mitochondrial division. Consequently, we focused our subsequent analyses on understanding the interaction between the WD-repeats of Net2p and Dnm1p.
Overexpression of Dnm1p not only rescues mitochondria from the effect of Net2WDp, it also changes the distribution of the Net2WD protein. When cells contained wild-type levels of Dnm1p, we observed that GFP-Net2WDp was evenly distributed in the cytosol (Figures 3B and 4B, left panel). In cells carrying both pGAL1-GFP-NET2WD and pGAL1-DNM1, several (5–15) small puncta of GFP-Net2WDp were seen in addition to faint cytosolic staining (Figure 4B, right panel). 3-D reconstruction of images from deconvolution microscopy showed that the dots of GFP-Net2WDp were not on the mitochondria (see supplemental data movie). Our observations therefore suggest that Dnm1p and Net2p interact via the WD-repeats of Net2p and that the Dnm1p-Net2p interaction can occur independent of mitochondria, thereby indicating that excess Dnm1p may titrate Net2WDp into complexes that can no longer act in a dominant negative manner. Supporting this view, we found that Dnm1p and GFP-Net2WDp interacted directly and could be coimmune precipitated from whole cell lysates (see Figure 6).
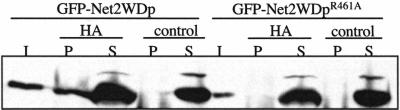
Figure 6.
Net2WDp, but not Net2WDpR461A, coimmune precipitates with Dnm1p-HA. dnm1Δ cells containing pHS15 (pGAL1-DNM1-HA) with either pKC68 (pGAL1-GFP-NET2WD) or pKC85 (pGAL1-GFP-NET2WDR461A) were lysed, and proteins were precipitated with HA antiserum. As a control, reactions were also incubated with protein A-sepharose alone. One hundred percent of supernatant (S) and pellet (P) fractions were analyzed by Western blotting, using GFP antibodies to detect GFP-Net2Np and GFP-Net2WDp. 30% of the total protein input (I) was used.
Mutations in the WD Repeats of Net2p Interfere with Net2p-Dnm1p Interaction
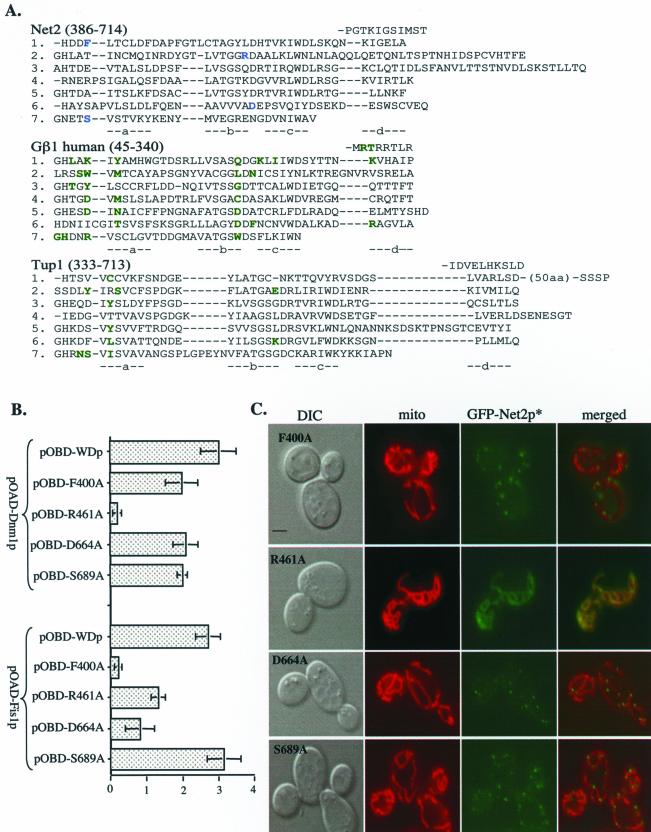
Structural analyses of several WD-repeat–containing proteins show that they all form similar shapes (Neer et al., 1994; Smith et al., 1999; Yu et al., 2000), and site-specific mutagenesis has identified residues that are important for protein-protein interactions (Wall et al., 1995; Sondek et al., 1996; Sprague et al., 2000). For example, amino acids located on the upper surface of the β-subunit of the heterotrimeric G protein, Gβ1, are required for binding to Gα and Gγ, not for the ability of Gβ1 to adopt its native conformation (see green residues in Figure 5A, and Gaudet et al., 1996; Sondek et al., 1996; Garcia-Higuera et al., 1998). Similar residues in Tup1p are critical for interaction with Matα2 (Komachi and Johnson, 1997; Sprague et al., 2000).
Figure 5.
Mutations in the WD-repeats of Net2p disrupt the Net2p-Dnm1p interaction. (A) Alignment of the WD-repeats of Net2p with Gβ1 and Tup1. Each WD repeat of the three proteins is listed on a separate line (1–7) and each β-strand of the propeller (a, b, c, d) is indicated below the protein sequences. The WD-repeats begin with the d strand of the 7th repeat, which is found immediately before the 1st WD repeat in the primary sequence. Amino acids of Net2p that were tested for interaction with Dnm1p are blue. Amino acids of Gβ1 that bind either Gγ and/or Gα are green, as are the residues of Tup1 that interact with Matα2. (B) Arginine 461 of Net2p is required for Dnm1p-Net2p interaction by two-hybrid β-galactosidase activities of cells with pOAD-Dnm1p and either pOBD-WDF400A, pOBD-WDR461A, pOBD-WDD664A, or pOBD-WDS689A are shown. The average of three independent experiments with one SD is shown. (C) Mutations in the WD-repeats impairs Net2p function and redistributes GFP-Net2p. net2Δ cells were transformed with pKC80, pKC81, pKC82, or pKC83, which express galactose-inducible fusions between GFP and Net2pF400A, Net2pR461A, Net2pD664A, or Net2pS689A, respectively. Localization of Net2p mutants was observed using DIC and fluorescence microscopy. Mitochondria were visualized by staining with mitofluor red 589. Bar, 3 μm.
On the basis of our alignment of Net2p with Tup1p and Gβ1, we altered residues predicted to be on the top face of the Net2WDp structure. Amino acids F400, R461, D664, and S689 of Net2WDp were changed to alanine and then tested for binding to Dnm1p and Fis1p using the yeast two-hybrid system. We found that three of our constructs, Net2WDpF400A, Net2WDpD664A, and Net2WDpS689A were only modestly altered in their association with Dnm1p in the two-hybrid assay (Figure 5B). In contrast, Net2WDpR461A was at least 20-fold reduced in its Dnm1p-interaction. Arguing that Net2WDpR461A was not grossly misfolded, we found that its binding to Fis1p was only mildly affected (Figure 5B). We found that the R461A mutation also altered the distribution of Net2p in yeast cells. Instead of the punctate location of wild-type GFP-Net2p, we found GFP-Net2pR461A evenly distributed along mitochondria (Figure 5C). Moreover, GFP-Net2pR461A did not rescue the mitochondrial fission defect of net2Δ cells nor did Net2WDpR461A act as a dominant negative in wild-type cells (K. Cerveny, unpublished results). Further supporting the role of R461 in Dnm1p binding, we found that Net2WDp, but not Net2WDpR461A, coimmune precipitated with Dnm1p. As shown in Figure 6, when Dnm1p-HA was precipitated from cells expressing Net2WDp, ∼30% of the Net2WDp was found in the pellet fraction along with Dnm1p-HA. In contrast, in cells expressing Dnm1p-HA and Net2WDpR461A, none of the Net2WDR461A protein precipitated with Dnm1p-HA.
Although mutations of residues D664 and S689 in the WD-repeats of Net2p did not seem to affect the interaction of Net2p with Dnm1p or Fis1p; the F400A mutation seemed to have the opposite effect as R461A. For instance, Net2WDpF400A is defective in its interaction with Fis1p, but not defective in Dnm1p-binding (Figure 5B). Also indicating that the Net2p-Dnm1p interaction is not disrupted by F400A, we found that GFP-Net2pF400A forms dot-like structures on mitochondria (Figure 5C). Net2pF400A only partially restores mitochondrial division in net2Δ cells (Figure 5C; 52% of cells contained partial networks or aggregated mitochondria, n = 80), and expression of Net2WDpF400A from the GAL1 promoter inhibits mitochondrial fission at a frequency similar to that of Net2WDp (74% of cells contained partial mitochondrial networks or aggregated mitochondria, n = 131). These data argue that the association of Net2WDp with both Fis1p and Dnm1p is important for Net2p function.
The GTPase Domain of Dnm1p Is Required for Interaction with Net2p
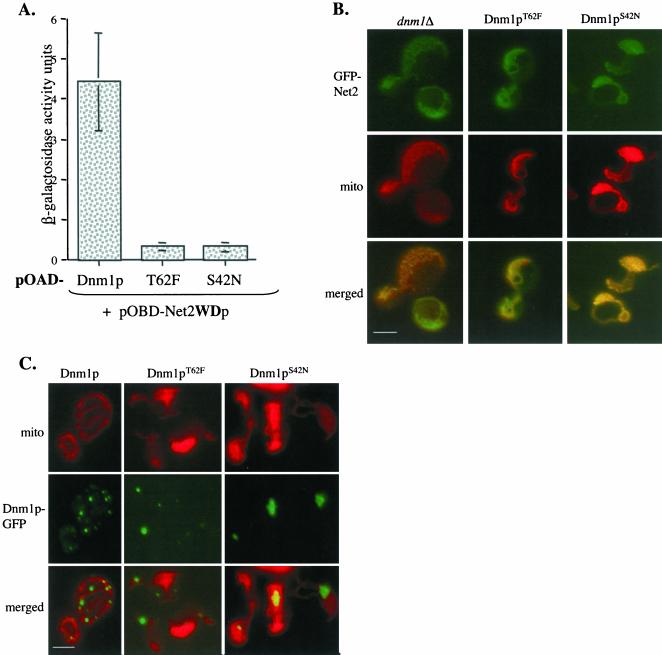
On the basis of mutations known to alter the GTPase activity of the homologous protein, dynamin (Damke et al., 2001; Hill et al., 2001), we made two corresponding changes in Dnm1p, S42N, and T62F. Constructs encoding Dnm1pS42N or Dnm1pT62F fused to the Gal4p activation domain were transformed into yeast cells expressing a Gal4p-DNA–binding domain-Net2WDp fusion. In contrast to cells with the wild-type Dnm1 protein, we found that neither of the altered Dnm1 proteins produced significant β-galactosidase activity (Figure 7A).
Figure 7.
Mutations in Dnm1p's GTPase region interfere with Net2p-Dnm1p interaction. (A) Dnm1p GTPase mutants do not interact with Net2p by two-hybrid β-galactosidase activities of cells with pOBD-WDp, and pOAD-Dnm1p, pOAD-T62F, or pOAD-S42N are shown. The averages of three separate experiments with error bars of one SD are shown. (B) Dnm1p GTPase activity is required to form Net2p puncta. dnm1Δ cells expressing GFP-Net2p from pKC62 in combination with either Dnm1S42N or Dnm1T62F from pKC78 or pKC79 were examined by fluorescence microscopy. Bar, 3 μm. (C) Dnm1p localization and distribution require an intact GTPase domain. WT cells expressing fusions between GFP and either Dnm1T62F or Dnm1S42N from plasmids pKC76 or pKC75 were examined using fluorescence microscopy. Bar, 3 μm.
An intact GTPase domain is also required for the normal distribution and function of Net2p. We expressed Dnm1p S42N or Dnm1p T62F in cells and examined the location of a GFP-Net2p fusion protein. In contrast to the punctate location of Net2p in wild-type cells, we found GFP-Net2p evenly distributed along the mitochondria in cells producing the mutated Dnm1 proteins (Figure 7B). This pattern of GFP-Net2p is similar to that seen in dnm1Δ disruption mutants and is consistent with our idea that binding and/or hydrolysis of GTP by Dnm1p is required for an interaction with Net2p. Consistent with previous studies (Otsuga et al., 1998), we found that alterations in Dnm1p's GTPase domain prevent mitochondrial division. A single, network of mitochondrial tubules was seen in the cells expressing Dnm1pS42N or Dnm1pT62F (Figure 7B). This defect was observed even in the presence of wild-type Dnm1p, indicating that these proteins block fission in a dominant-negative manner.
Wild-type Dnm1p appears to cycle between mitochondria and the cytosol. Cells carrying a Dnm1p-GFP fusion protein contain, on average, 25–30 dot-like structures on the mitochondria (Figure 7C). In addition, Dnm1p-GFP is found in numerous small, rapidly moving particles in the cytosol, some of which appear to assemble with the mitochondrial form of Dnm1p (K. Cerveny, H. Sesaki, and A. Aiken Hobbs, unpublished observations). We found that mutations in the GTPase region dramatically altered the normal pattern of Dnm1p. GFP fusions of Dnm1pS42N or Dnm1pT62F were found in four or five large aggregates on the mitochondria, and no cytosolic Dnm1p-GFP staining was visible (Figure 7C, and K. Cerveny, unpublished data).
Net2p and Fis1p Are Required for Normal Dnm1p Dynamics
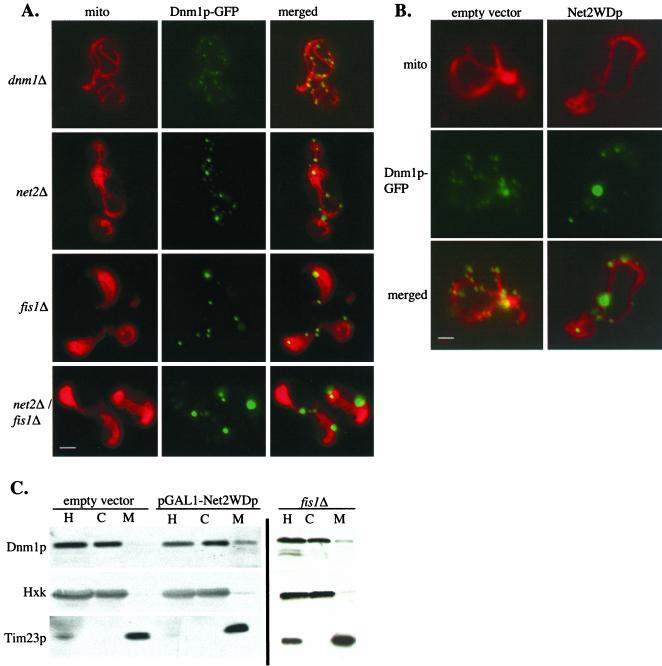
We next explored the requirement for Dnm1p's partner proteins, Net2p and Fis1p, in the distribution of Dnm1p between the mitochondria and cytosol. First, we examined Dnm1p-GFP in strains carrying net2Δ disruptions. In net2Δ cells, the pattern of Dnm1p-GFP was changed modestly. As shown in Figure 8A, instead of the 25–30 dot-like structures, each with a diameter of 300–400 nm on the mitochondria of wild-type cells, net2Δ cells contained fewer (∼15–18) and slightly larger (500–700 nm) Dnm1p-GFP particles on their mitochondria. The pattern of Dnm1p was more disrupted in cells expressing Net2WDp than in net2Δ cells. Galactose induction of cells carrying pKC68 (pGAL-NET2WD) caused a portion of Dnm1p-GFP to collapse into ∼2–3 large aggregates with fewer wild-type sized puncta. This distribution was similar to those seen in cells expressing Dnm1 proteins defective in their GTPase domain (compare Figure 8B with Figure 7C). Like cells expressing Dnm1pS42N or Dnm1pT62F, no trace of the cytosolic pool of Dnm1p-GFP could be detected. A similar, severe change in the distribution of Dnm1p-GFP was observed in fis1Δ and net2Δ fis1Δ cells (Figure 8A).
Figure 8.
Deletion of FIS1 or overproduction of Net2WDp causes Dnm1p to accumulate on mitochondria. (A) Lack of Fis1p changes the distribution of Dnm1p. dnm1Δ, net2Δ, fis1Δ, and net2Δ fis1Δ cells expressing Dnm1p-GFP from pHS20 were examined by fluorescence microscopy after staining mitochondria with mitofluor red 589. Bar, 3 μm. (B) Expression of Net2WDp causes Dnm1p to aggregate on mitochondria. Wild-type strain FY833 expressing Dnm1p-GFP from pHS20 and carrying either pKC68 (pGAL1-Net2WDp) or the empty vector pRS314GU were grown in galactose medium for 180 min. Cells were viewed by fluorescence microscopy. Bar, 3 μm. (C) A significant amount of Dnm1p cofractionates with mitochondria in either fis1Δ or Net2WDp-expressing cells. WT cells containing pRS316GU (empty vector) or pKC71 (pGAL1-Net2WDp) as well as fis1Δ cells with no plasmid were grown in galactose for 120 min. Cells were homogenized (H) and separated into a high-speed supernatant (C) and a mitochondrial pellet (M) by centrifugation. After SDS-PAGE, fractions were analyzed by Western blotting with antibodies to Dnm1p, Tim23p, and hexokinase (Hxk).
Subcellular fractionation data also supports our view that Dnm1p dynamics requires both Fis1p and Net2WDp. In wild-type cells, we found that the interaction of Dnm1p with mitochondria was labile, and very little Dnm1 protein cofractionated with mitochondria (Otsuga et al., 1998, Cerveny et al., 2001). Virtually all of Dnm1p was found in the cytosol (Figure 8C). However, a significant amount (∼10%) of Dnm1p was found on the mitochondria isolated from the fis1Δ mutant or from cells overproducing Net2WDp. To confirm that the sedimenting population of Dnm1p was associated with mitochondrial membranes and not simply aggregated, we solubilized mitochondria with nonionic detergents and found that Dnm1p was soluble (K. Cerveny, unpublished data). Our results thus indicate that defects in the GTPase domain of Dnm1p, the lack of Fis1p, and the overproduction of Net2WDp all cause Dnm1p to accumulate on the mitochondrial surface.
DISCUSSION
Our results show that the Net2 protein contains at least two functional domains, both playing important, but distinct, roles in mitochondrial division. Because the amino-terminal domain of Net2p interacts with Fis1p and the carboxyl-terminal region, Net2WDp, interacts with Dnm1p and because there is no evidence that Fis1p and Dnm1p directly interact (K. Cerveny, unpublished observations), it is tempting to speculate that Net2p links Fis1p and Dnm1p at sites of fission. However, our data suggest that Net2p is more than a simple bridging molecule. We show that the carboxyl-terminal WD-repeat region of Net2p interacts with both Dnm1p and Fis1p, and we have isolated mutations in the WD-repeat region of Net2p that primarily disrupt either Net2p-Dnm1p binding (Net2pR461A) or Net2p-Fis1p binding (Net2pF400A). Moreover, our data show that both of the mutated Net2 proteins are at least partially defective in mitochondrial division, indicating that the interaction of Net2p's WD-repeats with both Dnm1p and Fis1p is important for Net2p function.
Net2p is predicted to contain six or seven WD-40 repeats near its carboxyl-terminus. Structural studies have shown that the WD-40 repeats form seven blades, each consisting of four β-sheets connected by looped linker regions. These blades assemble into a barrel-shaped propeller structure that can act as a platform for protein-protein interactions (Wall et al., 1995; Saxena et al., 1996; Sondek et al., 1996; Sprague et al., 2000). In proteins such as Gβ1 and Tup1p, the top surface of the propeller contacts the partner proteins, with the loops between the β-strands providing much of the specificity (Garritsen and Simonds, 1994; Wall et al., 1995; Sondek et al., 1996; Komachi and Johnson, 1997; Sprague et al., 2000; Zhang et al., 2002). On the basis of our binding and localization analysis of the Net2p mutants, NET2F400A and NET2R461A, we propose that the WD-repeats of Net2p contact both Dnm1p and Fis1p on its top surface. The location of the analogous residues in the Tup1 structure, suggest that F400 and R461 may be close to each other (∼10 angstroms), raising the possibility that the Fis1p and Dnm1p binding sites on Net2WDp overlap.
Dnm1p is a dynamin-related GTPase, and the Net2p-Dnm1p association appears to depend on the nucleotide state of Dnm1p. Unlike wild-type Dnm1p, GTPase mutant versions of Dnm1p no longer interact with the WD-repeats of Net2p in the yeast two-hybrid assay. Moreover, in cells expressing the altered Dnm1 proteins, Net2p is no longer associated with Dnm1p in dot-like structures on the mitochondria. Instead, Net2p is evenly dispersed along the mitochondrial surface, presumably bound to the Fis1 protein. This distribution of Net2p is identical to that seen in dnm1Δ cells, arguing that the in vivo interaction of Net2p and Dnm1p is nucleotide dependent. Nearly all G-proteins undergo conformational changes upon GTP binding and hydrolysis, and many of these proteins such as Gα, ras, and rabs have been shown to preferentially interact with their partners in a nucleotide-dependent manner (Sarvazyan et al., 1998; Corbett and Alber, 2001; Li et al., 2001; Moyer et al., 2001; Short et al., 2001; Weide et al., 2001). Although the S42N and T62F mutations correspond to those already analyzed in the homologous dynamin protein, it remains controversial whether dynamin's GTP binding, hydrolysis, or both are affected by these changes (Hinshaw and Schmid, 1995; Binns et al., 1999; Sever et al., 1999, 2000; Damke et al., 2001; Marks et al., 2001; Eccleston et al., 2002). Furthermore, it is unclear how the enzymatic activities of Dnm1p and dynamin compare. Consequently, studies are underway to biochemically characterize purified versions of Dnm1p, Dnm1pS42N, and Dnm1pT62F.
Based on the properties of dynamin and its similarity to the Dnm1 protein, there are at least two possible models that explain the roles that Dnm1p, Net2p, and Fis1p play in mitochondrial division. Dynamin has been shown to form collars around clathrin-coated plasma membrane invaginations to mediate membrane scission (Takei et al., 1995). In one model for dynamin action, the assembly-stimulated GTP hydrolysis and the resulting conformational changes in the dynamin collar are proposed to constrict and pinch off the endocytic vesicle (Damke et al., 1994, 1995; Hinshaw and Schmid, 1995; Warnock et al., 1996; Sweitzer and Hinshaw, 1998). If Dnm1p acts in a similar manner, it could encircle mitochondria at specific sites and in a GTPase-dependent step, squeeze the mitochondrial tubule in two. In this scenario, Net2p and Fis1p would act primarily as scaffold proteins, anchoring Dnm1p to the mitochondria surface. In an alternative model, dynamin is proposed to function as a regulatory molecule that recruits and activates the actual membrane scission machinery in its GTP-bound state (Sever et al., 1999, 2000; Fish et al., 2000). If Dnm1p functions in this way, GTP-Dnm1p could either recruit factors, such as Net2p, to the site of scission, or activate proteins, like Fis1p, that may be already present.
Our data are most consistent with the idea that Dnm1p acts as a regulatory molecule, directly recruiting Net2p. For example, when the Net2p-Dnm1p interaction is disrupted, by either a mutation in the WD-repeats of Net2p or a defect in the GTPase region of Dnm1p, the distribution of Net2p changes dramatically. Instead of being in dot-like structures, the Net2 protein in cells expressing Net2pR461A, Dnm1pS42N, or Dnm1pT62F is found all along the mitochondrial surface. Dnm1p in these cells remains punctate. These observations argue that the location of Net2p is dependent on Dnm1p, but Dnm1p's distribution does not strictly require Net2p. Furthermore, if Net2p functions as a simple adapter molecule, bringing soluble Dnm1p together with the membrane-bound Fis1 protein, then excess Net2Np or Net2WDp would interfere with Net2p-Fis1p or Net2p-Dnm1p binding and cause release of Dnm1p from mitochondria. However, in cells overproducing Net2Np or Net2WDp, Dnm1p remains associated with mitochondria. Perhaps the strongest evidence that Dnm1p recruits Net2p is provided by our GAL1-NET2 induction studies. Net2p has two potential binding sites on mitochondria: either Fis1p, which is evenly distributed all along the outer membrane, or Dnm1p, which is located in discrete patches. When the expression of the Net2 protein is induced in cells, Net2p first appears in dot-like structures on mitochondria. Only when the Dnm1p-Net2p interaction is disrupted do we see Net2p all along the mitochondrial tubule bound to Fis1p. Together, our observations suggest that Dnm1p acts early in the division pathway and functions to recruit effector proteins, such as Net2p and Fis1p. Consistent with this idea, we find that Dnm1p can bind to specific sites on the mitochondria in the absence of either Fis1p or Net2p.
In a recent report, Tieu et al. (2002) suggest that Net2p functions as an adapter molecule, bringing together Fis1p and Dnm1p. Their data indicate that the amino-terminal region of Net2p interacts with Fis1p, whereas the carboxyl-terminal region binds only Dnm1p. They also showed that expression of either the amino- or carboxyl-terminal domains of Net2p block mitochondrial division. We confirmed and extended their studies by showing that Net2WDp was able to bind Dnm1p and, to a lesser extent, Fis1p. In addition, we identified key residues in the carboxyl-terminal WD-repeats of Net2p and the GTPase domain of Dnm1p that are required for assembly of the known mitochondrial fission components. Furthermore, we found that a direct physical interaction between Dnm1p and the WD-repeats of Net2p was responsible for the dominant inhibition of mitochondrial scission caused by Net2WDp. Tieu et al. (2002) proposed a model in which Fis1p recruits first Dnm1p and then Net2p to the site of mitochondrial division. However, because Fis1p is evenly distributed along the mitochondrial surface, it is difficult to imagine how Fis1p can recruit Dnm1p to a specific site. We suggest a different scenario, in which Dnm1p marks the site of fission and then recruits effector molecules, such as Net2p and Fis1p. Supporting this view, we see that much of Dnm1p remains on the mitochondria and in punctate spots in the absence of Fis1p. We speculate that some unknown protein or lipid modification is initially recognized by Dnm1p. Furthermore, although the precise role of Net2p in fission is not known, we suggest that at least one of the functions of Net2p is to coordinate the activities of Dnm1p and Fis1p by organizing the division complex. Consistent with this idea, we find that mutations in the GTPase region of Dnm1p, defects in the WD repeat-containing portion of Net2p, or the lack of Fis1p all seem to block mitochondrial fission at a common step. In all cases, Dnm1p is found in several large aggregates on the mitochondrial surface, and the cytosolic pool of Dnm1p appears to be absent. Additionally, if Dnm1p is artificially assembled (e.g., by fusion of dsRed to its carboxyl-terminus), mitochondrial scission is blocked and the dominant-negative Dnm1p-dsRed protein is found in large aggregates similar to those seen in Dnm1p and Net2p mutant cells (K. Cerveny, unpublished observations). Thus, the normal cycle of the Dnm1 protein appears to require productive and regulated interactions with Net2p, Fis1p, and Dnm1p, with Net2p playing a pivotal role in their coordination. Studies in progress are aimed at understanding the GTPase cycle of Dnm1p in the context of Net2p, Fis1p, and other potential mitochondrial fission proteins.
Supplementary Material
Acknowledgments
We thank M. Yaffe for F1β antisera, B. Glick for the RFP variant, S. Fields for pOAD and pOBD, and H. Sesaki for pHS78. We also thank C. Machamer, H. Sesaki, A. Aiken Hobbs, M. Youngman, C. Dunn, S. Southard, H. Hoard-Fruchey, C. Wolberger, and N. Clarke for productive discussions and critical comments on the manuscript. This work was supported by US Public Health Service Grant RO1-GM54021–06 to R.E.J.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E03–02–0092. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E03-02-0092.
Abbreviations used: GFP, green fluorescent protein; RFP and DsRed, red fluorescent protein; DIC, differential interference contrast; Sraf, synthetic medium with raffinose.
Online version of this article contains video and supplementary materials for some figures. Online version is available at www.molbiolcell.org.
References
- Adams, A., Gottschling, D., Kaiser, C., and Stearns, T. (1997). Methods in Yeast Genetics. Plainview, NY: Cold Spring Harbor Laboratory Press.
- Attardi, G., and Schatz, G. (1988). Biogenesis of mitochondria. Annu. Rev. Cell Biol. 4, 289–333. [DOI] [PubMed] [Google Scholar]
- Bai, C., and Elledge, S.J. (1996). Gene identification using the yeast two-hybrid system. Methods Enzymol. 273, 331–347. [DOI] [PubMed] [Google Scholar]
- Bereiter-Hahn, J., and Voth, M. (1994). Dynamics of mitochondria in living cells: shape changes, dislocations, fusion, and fission of mitochondria. Microsc. Res. Tech. 27, 198–219. [DOI] [PubMed] [Google Scholar]
- Binns, D.D., Barylko, B., Grichine, N., Atkinson, M.A., Helms, M.K., Jameson, D.M., Eccleston, J.F., and Albanesi, J.P. (1999). Correlation between self-association modes and GTPase activation of dynamin. J. Protein Chem. 18, 277–290. [DOI] [PubMed] [Google Scholar]
- Bleazard, W., McCaffery, J.M., King, E.J., Bale, S., Mozdy, A., Tieu, Q., Nunnari, J., and Shaw, J.M. (1999). The dynamin-related GTPase Dnm1 regulates mitochondrial fission in yeast. Nat. Cell Biol. 1, 298–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerveny, K.L., McCaffery, J.M., and Jensen, R.E. (2001). Division of mitochondria requires a novel DMN1-interacting protein, Net2p. Mol. Biol. Cell 12, 309–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbett, K.D., and Alber, T. (2001). The many faces of Ras: recognition of small GTP-binding proteins. Trends Biochem. Sci. 26, 710–716. [DOI] [PubMed] [Google Scholar]
- Damke, H., Baba, T., Warnock, D.E., and Schmid, S.L. (1994). Induction of mutant dynamin specifically blocks endocytic coated vesicle formation. J. Cell Biol. 127, 915–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damke, H., Binns, D.D., Ueda, H., Schmid, S.L., and Baba, T. (2001). Dynamin GTPase domain mutants block endocytic vesicle formation at morphologically distinct stages. Mol. Biol. Cell 12, 2578–2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damke, H., Gossen, M., Freundlieb, S., Bujard, H., and Schmid, S.L. (1995). Tightly regulated and inducible expression of dominant interfering dynamin mutant in stably transformed HeLa cells. Methods Enzymol. 257, 209–220. [DOI] [PubMed] [Google Scholar]
- Daum, G., Böhni, P.C., and Schatz, G. (1982). Import of proteins into mitochondria. Cytochrome b2 and cytochrome c peroxidase are located in the intermembrane space of yeast mitochondria. J. Cell Biol. 257, 13028–13033. [PubMed] [Google Scholar]
- Davis, A.J., Ryan, K.R., and Jensen, R.E. (1998). Tim23p contains separate and distinct signals for targeting to mitochondria and insertion into the inner membrane. Mol. Biol. Cell 9, 2577–2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eccleston, J.F., Binns, D.D., Davis, C.T., Albanesi, J.P., and Jameson, D.M. (2002). Oligomerization and kinetic mechanism of the dynamin GTPase. Eur. Biophys. J. 31, 275–282. [DOI] [PubMed] [Google Scholar]
- Emtage, J.L., and Jensen, R.E. (1993). MAS6 encodes an essential inner membrane component of the yeast mitochondrial protein import pathway. J. Cell Biol. 122, 1003–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fekkes, P., Shepard, K.A., and Yaffe, M.P. (2000). Gag3p, an outer membrane protein required for fission of mitochondrial tubules. J. Cell Biol. 151, 333–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fish, K.N., Schmid, S.L., and Damke, H. (2000). Evidence that dynamin-2 functions as a signal-transducing GTPase. J. Cell Biol. 150, 145–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukushima, N.H., Brisch, E., Keegan, B.R., Bleazard, W., and Shaw, J.M. (2001). The GTPase effector domain sequence of the Dnm1p GTPase regulates self-assembly and controls a rate-limiting step in mitochondrial fission. Mol. Biol. Cell 12, 2756–2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gammie, A.E., Kurihara, L.J., Vallee, R.B., and Rose, M.D. (1995). DNM1, a dynamin-related gene, participates in endosomal trafficking in yeast. J. Cell Biol. 130, 553–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Higuera, I., Gaitatzes, C., Smith, T.F., and Neer, E.J. (1998). Folding a WD repeat propeller. Role of highly conserved aspartic acid residues in the G protein beta subunit and Sec13. J. Biol. Chem. 273, 9041–9049. [DOI] [PubMed] [Google Scholar]
- Garritsen, A., and Simonds, W.F. (1994). Multiple domains of G protein beta confer subunit specificity in beta gamma interaction. J. Biol. Chem. 269, 24418–24423. [PubMed] [Google Scholar]
- Gaudet, R., Bohm, A., and Sigler, P.B. (1996). Crystal structure at 2.4 angstroms resolution of the complex of transducin betagamma and its regulator, phosducin. Cell 87, 577–588. [DOI] [PubMed] [Google Scholar]
- Gottlieb, R.A. (2000). Mitochondria: execution central. FEBS Lett. 482, 6–12. [DOI] [PubMed] [Google Scholar]
- Haid, A., and Suissa, M. (1983). Immunochemical identification of membrane proteins after sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Methods Enzymol. 96, 192–205. [DOI] [PubMed] [Google Scholar]
- Hales, K.G., and Fuller, M.T. (1997). Developmentally regulated mitochondrial fusion mediated by a conserved, novel, predicted GTPase. Cell 90, 121–129. [DOI] [PubMed] [Google Scholar]
- Harlow, E., and Lane, D. (1988). Antibodies—A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
- Hermann, G.J., and Shaw, J.M. (1998). Mitochondrial dynamics in yeast. Annu. Rev. Cell Dev. Biol. 14, 265–303. [DOI] [PubMed] [Google Scholar]
- Hill, E., van Der Kaay, J., Downes, C.P., and Smythe, E. (2001). The role of dynamin and its binding partners in coated pit invagination and scission. J. Cell Biol. 152, 309–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinshaw, J.E., and Schmid, S.L. (1995). Dynamin self-assembles into rings suggesting a mechanism for coated vesicle budding. Nature 374, 190–192. [DOI] [PubMed] [Google Scholar]
- Jensen, R.E., Hobbs, A.E., Cerveny, K.L., and Sesaki, H. (2000). Yeast mitochondrial dynamics: fusion, division, segregation, and shape. Microsc. Res. Tech. 51, 573–583. [DOI] [PubMed] [Google Scholar]
- Komachi, K., and Johnson, A.D. (1997). Residues in the WD repeats of Tup1 required for interaction with alpha2. Mol. Cell. Biol. 17, 6023–6028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labrousse, A.M., Zappaterrs, M.D., Rube, D.A., and van der Bliek, A.M. (1999). C. elegans dynamin-related protein DRP-1 controls severing of the mitochondrial outer membrane. Mol. Cell 4, 815–826. [DOI] [PubMed] [Google Scholar]
- Laemmli, U.K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685. [DOI] [PubMed] [Google Scholar]
- Li, W., Chong, H., and Guan, K.L. (2001). Function of the Rho family GTPases in Ras-stimulated Raf activation. J. Biol. Chem. 276, 34728–34737. [DOI] [PubMed] [Google Scholar]
- Marks, B., Stowell, M.H., Vallis, Y., Mills, I.G., Gibson, A., Hopkins, C.R., and McMahon, H.T. (2001). GTPase activity of dynamin and resulting conformation change are essential for endocytosis. Nature 410, 231–235. [DOI] [PubMed] [Google Scholar]
- Miyakawa, I., Aoi, H., Sando, N., and Kuroiwa, T. (1984). Fluorescence microscopic studies of mitochondrial nucleoids during meiosis and sporulation in the yeast, Saccharomyces cerevisiae. J. Cell Sci. 66, 21–38. [DOI] [PubMed] [Google Scholar]
- Moyer, B.D., Allan, B.B., and Balch, W.E. (2001). Rab1 interaction with a GM130 effector complex regulates COPII vesicle cis-Golgi tethering. Traffic 2, 268–276. [DOI] [PubMed] [Google Scholar]
- Mozdy, A.D., McCaffery, J.M., and Shaw, J.M. (2000). Dnm1p GTPase-mediated mitochondrial fission Is a multi-step process requiring the novel integral membrane component Fis1p. J. Cell Biol. 151, 367–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neer, E.J., Schmidt, C.J., Nambudripad, R., and Smith, T.F. (1994). The ancient regulatory-protein family of WD-repeat proteins. Nature 371, 297–300. [DOI] [PubMed] [Google Scholar]
- Nieman, H.L., Houghten, R.A., Walker, L.E., Reisfeld, R.A., Wilson, I.A., Hogle, J.M., Lerner, R.A. (1983). Generation of protein-reactive antibodies by short peptides is an event of high frequency: implications for the structural basis of immune recognition. Proc. Natl. Acad. Sci. USA 80, 4949–4953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsuga, D., Keegan, B.R., Brisch, E., Thatcher, J.W., Hermann, G.J., Bleazard, W., and Shaw, J.M. (1998). The dynamin-related GTPase, Dnm1p, controls mitochondrial morphology in yeast. J. Cell Biol. 143, 333–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarvazyan, N.A., Remmers, A.E., and Neubig, R.R. (1998). Determinants of gi1alpha and beta gamma binding. Measuring high affinity interactions in a lipid environment using flow cytometry. J. Biol. Chem. 273, 7934–7940. [DOI] [PubMed] [Google Scholar]
- Saxena, K., Gaitatzes, C., Walsh, M.T., Eck, M., Neer, E.J., and Smith, T.F. (1996). Analysis of the physical properties and molecular modeling of Sec 13, A WD repeat protein involved in vesicular traffic. Biochemistry 35, 15215–15221. [DOI] [PubMed] [Google Scholar]
- Sesaki, H., and Jensen, R.E. (1999). Division versus fusion: Dnm1p and Fzo1p antagonistically regulate mitochondrial shape. J. Cell Biol. 147, 699–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sesaki, H., and Jensen, R.E. (2001). UGO1 encodes an outer membrane protein required for mitochondrial fusion. J. Cell Biol. 152, 1123–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sever, S., Damke, H., and Schmid, S.L. (2000). Dynamin:GTP controls the formation of constricted coated pits, the rate limiting step in clathrin-mediated endocytosis. J. Cell Biol. 150, 1137–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sever, S., Muhlberg, A.B., and Schmid, S.L. (1999). Impairment of dynamin's GAP domain stimulates receptor-mediated endocytosis. Nature 398, 481–486. [DOI] [PubMed] [Google Scholar]
- Short, B., Preisinger, C., Korner, R., Kopajtich, R., Byron, O., and Barr, F.A. (2001). A GRASP55-rab2 effector complex linking Golgi structure to membrane traffic. J. Cell Biol. 155, 877–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski, R.S., and Hieter, P. (1989). A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122, 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, T.F., Gaitatzes, C., Saxena, K., and Neer, E.J. (1999). The WD repeat: a common architecture for diverse functions. Trends Biochem. Sci. 24, 181–185. [DOI] [PubMed] [Google Scholar]
- Sondek, J., Bohm, A., Lambright, D.G., Hamm, H.E., and Sigler, P.B. (1996). Crystal structure of a G-protein beta gamma dimer at 2.1A resolution. Nature 379, 369–374. [DOI] [PubMed] [Google Scholar]
- Sprague, E.R., Redd, M.J., Johnson, A.D., and Wolberger, C. (2000). Structure of the C-terminal domain of Tup1, a corepressor of transcription in yeast. EMBO J. 19, 3016–3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweitzer, S.M., and Hinshaw, J.E. (1998). Dynamin undergoes a GTP-dependent conformational change causing vesiculation. Cell 93, 1021–1029. [DOI] [PubMed] [Google Scholar]
- Takei, K., McPherson, P.S., Schmid, S.L., and De Camilli, P. (1995). Tubular membrane invaginations coated by dynamin rings are induced by GTP-gamma S in nerve terminals. Nature 374, 186–190. [DOI] [PubMed] [Google Scholar]
- Tieu, Q., and Nunnari, J. (2000). Mdv1p is a WD repeat protein that interacts with the dynamin-related GTPase, Dnm1p, to trigger mitochondrial division. J. Cell Biol. 151, 353–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tieu, Q., Okreglak, V., Naylor, K., and Nunnari, J. (2002). The WD repeat protein, Mdv1p, functions as a molecular adaptor by interacting with Dnm1p and Fis1p during mitochondrial fission. J. Cell Biol. 158, 445–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzagoloff, A. (1983). Mitochondria. New York: Plenum Press.
- Uetz, P. et al. (2000). A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature 403, 623–627. [DOI] [PubMed] [Google Scholar]
- Wall, M.A., Coleman, D.E., Lee, E., Iniguez-Lluhi, J.A., Posner, B.A., Gilman, A.G., and Sprang, S.R. (1995). The structure of the G protein heterotrimer Gi alpha 1 beta 1 gamma 2. Cell 83, 1047–1058. [DOI] [PubMed] [Google Scholar]
- Warnock, D.E., Hinshaw, J.E., and Schmid, S.L. (1996). Dynamin self-assembly stimulates its GTPase activity. J. Biol. Chem. 271, 22310–22314. [DOI] [PubMed] [Google Scholar]
- Weide, T., Bayer, M., Koster, M., Siebrasse, J.P., Peters, R., and Barnekow, A. (2001). The Golgi matrix protein GM 130, a specific interacting partner of the small GTPase rab1b. EMBO Rep. 2, 336–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winston, F., Dollard, C., and Ricupero-Hovasse, S.L. (1995). Construction of a set of convenient Saccharomyces cerevisiae strains that are isogenic to S288C. Yeast 11, 53–55. [DOI] [PubMed] [Google Scholar]
- Yaffe, M.P., and Schatz, G. (1984). Two nuclear mutations that block mitochondrial protein import in yeast. Proc. Natl. Acad. Sci. USA 81, 4819–4823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, L., Gaitatzes, C., Neer, E., and Smith, T.F. (2000). Thirty-plus functional families from a single motif. Protein Sci. 9, 2470–2476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Z., Varanasi, U., Carrico, P., and Trumbly, R.J. (2002). Mutations of the WD repeats that compromise Tup1 repression function maintain structural integrity of the WD domain trypsin-resistant core. Arch. Biochem. Biophys. 406, 47–54. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.