Abstract
Bass Myo3A, a class III myosin, was expressed in HeLa cells as a GFP fusion in order to study its cellular localization. GFP-Myo3A localized to the cytoplasm and to the tips of F-actin bundles in filopodia, a localization that is consistent with the observed concentration toward the distal ends of F-actin bundles in photoreceptor cells. A mutation in the motor active site resulted in a loss of filopodia localization, suggesting that Myo3A motor activity is required for filopodial tip localization. Deletion analyses showed that the NH2-terminal kinase domain is not required but the CO2H-terminal 22 amino acids of the Myo3A tail are required for filopodial localization. Expression of this tail fragment alone produced fluorescence associated with F-actin throughout the cytoplasm and filopodia and a recombinant tail fragment bound to F-actin in vitro. An actin-binding motif was identified within this tail fragment, and a mutation within this motif abolished both filopodia localization by Myo3A and F-actin binding by the tail fragment alone. Calmodulin localized to filopodial tips when coexpressed with Myo3A but not in the absence of Myo3A, an observation consistent with the previous proposal that class III myosins bind calmodulin and thereby localize it in certain cell types.
INTRODUCTION
Myosins are motor proteins that use ATP hydrolysis to translocate along actin filaments and thereby provide the power for muscle contraction and many other cellular movements, including directed molecular transport and organelle localization. Numerous types of myosins have been discovered, and based on the primary structure of their motor domain, they are classified into at least 18 classes (Hodge and Cope, 2000).
The primary structure of myosins includes an N-terminal motor domain of ∼80 kDa that contains nucleotide- and actin-binding sites followed by a short, regulatory neck domain containing IQ motifs that interact with calmodulin and/or light chains and a structurally unique, C-terminal tail domain that mediates class-specific function. Class III myosins are distinctive in also having a kinase domain N-terminal to the motor domain. The vertebrate class III myosins have one or more IQ motifs present in the tail region beyond the usual neck domain location.
Class III myosins are associated with sensory systems in both vertebrates and invertebrates. The ninaC gene from D. melanogaster expresses two class III myosins from the same gene via differential mRNA splicing (Montell and Rubin, 1988). Null mutations in ninaC delay termination of the photoreceptor response and lead to progressive retinal degeneration (Porter et al., 1993). Four vertebrate class III myosin genes have been identified: two from human, MYO3A and MYO3B (Dosé and Burnside, 2000, 2002), and two from striped bass, Myo3A and Myo3B (Dosé et al., 2003). Expression of vertebrate myosin IIIA is restricted, with the strongest expression in retina and cochlea (Dosé and Burnside, 2000; Dosé et al., 2003; Walsh et al., 2002). Normal hearing in humans involves myosin IIIA as mutations in MYO3A causes nonsyndromic progressive hearing loss (Walsh et al., 2002). Immunolocalization studies have shown that striped bass Myo3A localizes to photoreceptors, specifically at the distal ends of long, ellipsoidal actin bundles that terminate in the calycal processes (Dosé et al., 2003), filopodia-like cellular extensions of unknown function.
The kinase domains of class III myosins show considerable sequence homology to Ste20/PAK family of protein kinases (Dan et al., 2001; Dosé et al., 2003), some of which function in signaling pathways that directly regulate myosin motor activity. Both human myosin III and Drosophila NINAC kinase domains have protein kinase activity in vitro and undergo autophosphorylation where the sites map to the motor domain (Ng et al., 1996; Komaba et al., 2003). Transgenic studies in which ninaC mutants are expressed in a ninaC-null background have revealed that the kinase domain is required for a normal electrophysiological photoresponse (Porter and Montell, 1993).
Myosin tail domains often mediate class-specific functions by dictating specific cellular localization or by binding to specific cargo molecules. Little is known about the functions of class III myosin tails except in Drosophila, where the tail domains of the two NINAC isoforms each confer a distinct subcellular localization within the photoreceptor (Porter et al., 1992). Overall, class III myosin tails show few sequence similarities except for two highly conserved motifs located near the extreme C-terminus of the vertebrate myosin III proteins. These motifs, termed myosin III tail homology domain I (3THDI) and myosin III tail homology domain II (3THDII; Dosé et al., 2003), likely serve important functions. Myosin 3THDII is present in both myosin IIIA and IIIB, whereas 3THDII is unique to myosin IIIA proteins.
Here, we ectopically expressed striped bass Myo3A in HeLa cells in order to study its cellular localization. Cells expressing a green fluorescent protein (GFP)-Myo3A fusion showed a striking localization of fluorescence at the distal tips of actin bundles in filopodia. This observation was used to analyze the role of specific Myo3A domains in filopodial localization and to determine if Myo3A colocalizes with calmodulin.
MATERIALS AND METHODS
GFP Expression Gene Constructs
Striped bass Myo3A gene fragments were produced using gene specific DNA primers (Table 1) and Myo3A cDNA template (Dosé et al., 2003; GenBank accession number AF003249) in polymerase chain reactions (PCR). All Myo3A fragments were engineered to have a 5′-end EcoRI site and a 3′-end SalI restriction sites for cloning into expression vectors. PCR fragments were initially cloned using the pCRII-TOPO cloning kit (Invitrogen, San Diego, CA), and all constructs were verified by DNA sequencing. Cloned fragments were excised and ligated into the EcoRI and SalI sites of either pEGFP-C2 or pEGFP-C3 expression vectors (Clontech, Palo Alto, CA) for expression in HeLa cells. For production of recombinant proteins, Myo3A fragments were inserted into the EcoRI and SalI restriction sites of pGEX-4T1 vector (Pharmacia, Piscataway, NJ). Escherichia coli strain BL21 was used for protein expression. GFP:M3, GFP:M3Δ3THDIΔ3THDII, GFP:M3ΔTHDII, and GFP:M3-inactive constructs have a c-Myc epitope tag sequence (EQKLISEEDL) located between the start codon and the second codon of the Myo3A reading frame. GFP:3THDI,3THDII contains a FLAG epitope sequence (DYKDDDDK) just upstream of the Myo3A sequences. Site-directed alanine mutations were constructed using two-step PCR and primers containing the altered codons (changed nucleotides are underlined in primer sequence in Table 1). Primers used to make deletion and truncation fragments contain start or stop codons where required. For the untagged version of Myo3A, the Myo3A sequence from GFP:M3 was inserted in the EcoRI and SalI sites of pEGFP-N1 vector (Clontech), which places the GFP sequences downstream of the Myo3A stop codon. GFP:CaM contains the human calmodulin cDNA engineered to have a 5′-end SalI site just upstream of the start codon and a 3′-end NotI site after the stop codon; this DNA fragment was cloned into the SalI and NotI sites of pFastBac plasmid (Invitrogen), from which the CaM fragment was excised as a SalI/XhoI fragment and cloned into the SalI site of pEGFP-C3 vector.
Table 1.
Primers used in this study
| DNA Construct (amino acids) | 5′-end PCR primer | 3′-end PCR primer |
|---|---|---|
| GFP:M3 (1-1838) | 5′-cgaattcaaacaatggaacaaaaactcatctca- | 5′-aagtcgactcagtactgtttgatgagcttccg-3′ |
| gaagaggatctgagatctctcctgaagatgtttc-3′ | ||
| GFP:M3-inactive | 5′-aatgacaactccagcgcctttggcaagtacctg-3′ | 5′-tcctcatattcaatcatc-3′ |
| (1-1838) step 1 | ||
| GFP:M3-inactive | 5′-ctagaagttctagacgag-3′ | Product from step 1 |
| (1-1838) step 2 | ||
| GFP:M3Δkinase | 5′-aagaattcggtgtggacgacctcgccacccta-3′ | 5′-aagtcgactcagtactgtttgatgagcttccg-3′ |
| (361-1838) | ||
| GFP:M3 | 5′-cgaattcaaacaatggaacaaaaactcat- | 5′-aagtcgactcattgatgggcctggatgatctc-3′ |
| Δ3THDIΔ3THDII | ctcagaagaggatctgagatctctcctgaagat- | |
| (1-1662) | gtttc-3′ | |
| GFP:M3Δ3THDII | 5′-cgaattcaaacaatggaacaaaaactcatctca- | 5′-aagtcgactcagtcgtcagtatggctctc-3′ |
| (1-1816) | gaagaggatctgagatctctcctgaagatgtttc-3′ | |
| GFP:3THDI,3THDII | 5′-agaattcaaacaatggactacaaggatgacgat- | 5′-aagtcgactcagtactgtttgatgagcttccg-3′ |
| (1620-1838) | gacaaggccaacaagcaaacagcgcag-3′ | |
| GFP:3THDII (1817-1838) | 5′-aagaattcggtaatccgtacgacttcagacat-3′ | 5′-aagtcgactcagtactgtttgatgagcttccg-3′ |
| GFP:3THDI (1620-1816) | 5′-aagaattcggtaatccgtacgacttcagacat-3′ | 5′-aagtcgactcagtcgtcagtatggctctc-3′ |
| GFP:M3 R/A (1-1838) | 5′-cgaattcaaacaatggaacaaaaactcatctca- | 5′-aagtcgactcagtactgtttgatgagcttccgtctctgggaggtcttcct- |
| gaagaggatctgagatctctcctgaagatgtttc-3′ | cagtagatgtgcgaagtcgtacgga-3′ | |
| GFP:3THDII R/A | 5′-aagaattcggtaatccgtacgacttcagacat-3′ | 5′-aagtcgactcagtactgtttgatgagcttccgtctctgggaggtcttcctcag- |
| (1817-1838) | tagatgtgcgaagtcgtacgga-3′ | |
| GFP:CaM | 5′-cgtcgaccaccatggctgaccaactgactgaa-3′ | 5′-cgtagcggccgctcactttgctgtcatcatttgtacaaa-3′ |
Bases involved in site-directed mutagenesis are underlined.
Cell Culture and Transfections
HeLa cells were maintained in DMEM + 10% fetal bovine serum. Cells were grown in six-well dishes containing poly-lysine–treated glass cover slips and transfected with 2 μg of plasmid DNA using 6 μl of FuGene 6 transfection reagent (Roche, Indianapolis, IN), according to the manufacturer's instructions. Twenty to twenty-four hours after transfection, the cells were fixed with 4% paraformaldehyde in phosphate-buffered saline (PBS) at 4°C for 30 min and permeabilized with 0.1% Triton-X100 in PBS for 10 min. Permeabilized cells were stained with Texas Red–phalloidin (Molecular Probes, Eugene, OR) used at 1:1000 dilution in PBS to visualize the actin cytoskeleton. All transfections and localization analyses were repeated a minimum of three times to confirm results.
Imaging and Analysis
Cells were observed with an inverted microscope (Carl Zeiss, Inc., Thornwood, NY) using a 63×/NA 1.4 objective. Images were acquired with a cooled CCD camera (Hamamatsu Corp., Bridgewater, NJ) controlled by Open Lab software (Improvision Inc., Lexington, MA) and processed and labeled using Adobe Photoshop software (San Jose, CA).
Immunolocalization of Myo3A in HeLa Cells
HeLa cells were fixed and F-actin stained as above and incubated with a 1:1000 dilution of rabbit polyclonal anti-Myo3A antibody (Dosé et al., 2003). The primary antibody was visualized using goat anti-rabbit Alexa 488 secondary antibody. Images were acquired and processed as above. The primary antibody weakly stained (cross-reacted) with the nucleus of HeLa cells, independent of Myo3A expression.
Actin Spin-down Assay
Recombinant GST proteins were expressed from pGEX (Pharmacia) vectors in BL21[DE3] E. coli cell line and purified using glutathione-agarose beads (Sigma, St. Louis, MO) per manufacturers protocol. Actin filaments were formed in vitro from mammalian actin monomers as described (Goode et al., 1999). Nonmuscle actin (1 mg/ml; Cytoskeleton, Inc., Boulder, CO) was polymerized for 1 h at room temperature in 50 mM KCl, 2 mM MgCl2, 1 mM ATP. Purified GST-fusion proteins were prespun for 1 h 24°C at 150,000 × g to remove aggregated protein. The prespun protein was incubated with F-actin for 30 min in 5 mM Tris-HCl, pH 8.0, 43 mM KCl, 0.02 mM CaCl2, 2 mM MgCl2,1 mM ATP. Fifty microliters of this reaction was layered onto 100 μl of F-actin cushion buffer (5 mM Tris-HCl, pH 8.0, 50 mM KCl, 2 mM MgCl2, 10% glycerol [vol/vol]) and centrifuged for 1.5 h at 24°C at 150,000 × g. Thirty microliters of supernatant was removed for analysis using SDS-PAGE, and the remaining supernatant including the cushion was removed and the sides of the tube and pellet were washed with 5 mM Tris-HCl, pH 8.0, 0.2 mM CaCl2, 0.2 mM ATP. The pellet was dissolved in 50 μl SDS sample buffer and three-fifths of it was taken to compare to the supernatant fraction by SDS-PAGE. Concentrations of fusion proteins and F-actin in the reactions were as follows: F-actin 9.5 μM, GST 8.2 μM, GST:3THDII 6.1 μM, GST:3THDII R/A 5.8 μM.
RESULTS
GFP-Myo3A Localizes to Filopodia Tips in Transfected HeLa Cells
HeLa cells were transfected with a series of GFP-Myo3A constructs (Figure 1) and analyzed by fluorescent microscopy. Representative composite images of cells showing GFP fluorescence (green) and F-actin staining (red) are shown in Figure 2. Control cells expressing GFP alone showed fluorescence throughout the cell body, including the nucleus (Figure 2A). Cells expressing a full-length Myo3A fusion (GFP:M3) exhibited diffuse fluorescence throughout the cytoplasm, with most cells (86%, n = 100 transfected cells) also showing concentrated staining in the tips of actin filament bundles that form the core of cellular projections (Figure 2B). No nuclear fluorescence was observed. It appears that these labeled projections are mostly filopodia rather than retraction fibers, because similar projections formed within 1 to 2 h after cell reattachment in replating assays, and GFP:M3 was found again highly concentrated at the tips of these newly formed projections (our unpublished results). GFP:M3 did not appear to strongly colocalize with other F-actin present in these cells, such as the filaments located in stress fibers or in the cortical meshwork at the cell periphery. Expression of GFP:M3 in either COS7 cells or NG-108 neuroblastoma-glioma cells produced a pattern of fluorescence similar to that in HeLa cells: i.e., both diffuse cytoplasmic fluorescence and concentrated fluorescence at the tips of F-actin–filled projections (our unpublished results).
Figure 1.
Schematic diagrams of striped bass Myo3A-GFP fusion constructs with the corresponding amino acids present in each construct shown in parentheses.
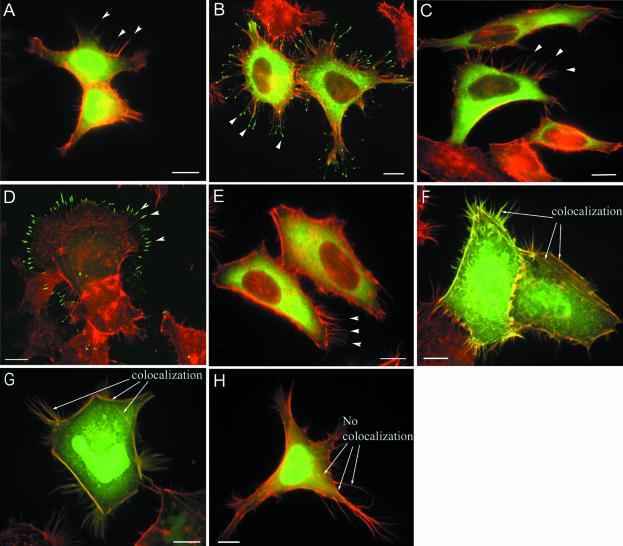
Figure 2.
Localization of GFP-myosin IIIA proteins in transfected HeLa cells. Composite images of GFP fluorescence (green) and F-actin staining (red) are shown. (A) GFP localizes throughout the cytoplasm and nucleus but not to filopodial tips. (B) GFP:M3 localizes to the cytoplasm and filopodial tips. (C) GFP:M3-inactive localizes to the cytoplasm but not filopodial tips. (D) GFP:M3Δkinase localizes to the cytoplasm and filopodial tips. (E) GFP:M3Δ3THDII localizes to the cytoplasm but not to filopodial tips. (F) GFP:3THDI,3THDII localizes throughout the cell and colocalizes with actin filaments (as seen by yellow staining) in the cell body, cortex, and protrusions. (G) GFP:3THDII localizes throughout the cell and colocalizes with actin filaments. (H) GFP:3THDI localizes throughout the cell but not with actin structures. Arrowheads point to representative filopodial tips; arrows point to examples of F-actin-containing structures that show colocalization with GFP (F and G) or no localization with GFP (H); bars, 10 μm.
To determine whether filopodia tip localization of Myo3A depends on motor activity, a construct, GFP:M3-inactive, was generated that harbored a point mutation that should render the Myo3A motor inactive. The codon for arginine 507 in the motor active site was altered to encode alanine. This arginine is strictly conserved in myosin motor domains and was shown to be essential for ATP-cleaving activity and motility in Dictyostelium discoideum myosin I (Shimada et al., 1997). Cells expressing GFP:M3-inactive showed diffuse cytoplasmic fluorescence and no apparent fluorescence in filopodia (Figure 2C), which implies that motor activity is required for localization of Myo3A to filopodia tips.
To determine the role of the Myo3A kinase domain in filopodial tip localization, a construct lacking the complete kinase domain (GFP:M3Δkinase) was generated. Cells expressing GFP:M3Δkinase showed both weak cytoplasmic fluorescence and concentrated fluorescence at filopodia tips in 95% of the transfected cells (n = 100) (Figure 2D), which indicates the kinase domain is not required for Myo3A tip localization. Interestingly, most transfected cells exhibited enhanced filopodia tip fluorescence and reduced cytoplasmic fluorescence compared with that seen in cells expressing full-length GFP-Myo3A (compare B with D in Figure 2).
To determine the role of the Myo3A tail domain in tip localization, two different CO2H-terminal truncation constructs were generated: GFP:M3Δ3THDI,Δ3THDII, which lacks the last 176 amino acids of Myo3A including both conserved tail domains, 3THDI and 3THDII; and GFP: M3Δ3THDII, which lacks the last 22 amino acids of the tail, the 3THDII. Cells expressing either tail truncation construct showed only diffuse cytoplasmic fluorescence with no apparent fluorescence in filopodia (Figure 2E; GFP: M3Δ3THDIΔ3THDII our unpublished results), which indicates the 3THDII must be present for filopodial tip localization by Myo3A.
To determine whether Myo3A tail sequences alone could localize to filopodia tips, GFP fusions consisting of Myo3A tail fragments, specifically the portions deleted in the above tail truncation constructs, were generated. Cells expressing either GFP:3THDI,3THDII (Figure 2F) or GFP:3THDII (Figure 2G) produced fluorescence throughout the cell and showed specific labeling of fibrous structures in the cell body, the cortical region at the cell periphery, and the entire lengths of cell protrusions. These labeled structures and sites also stained with Texas Red-phalloidin, suggesting that these tail fragments are colocalizing with actin filaments in stress fibers, the cell cortex, and cellular projections. The ability of GFP:3THDII to colocalize with actin filaments suggests that 3THDII binds actin and is likely responsible for the observed actin filament association by the larger tail fragment. In agreement with this conclusion, expression of GFP:3THDI, a tail construct lacking 3THDII, did not show actin filament costaining (Figure 2H).
Point Mutation in Putative Actin-binding Motif Abolishes Filopodial Tip Localization
The 22-amino acid sequence of 3TDHII is highly conserved between fish and human myosin IIIA tails (Figure 3A). On reviewing the scientific literature on known actin-binding motifs, we discovered that the amino acid sequence DFRXXL (where X denotes any amino acid) is present within the 3TDHII of both fish and human myosin IIIA (Figure 3A). The DFRXXL sequence was identified recently as a novel actin-binding motif in smooth muscle myosin light-chain kinase (MLCK; Smith et al., 1999). Three copies of this motif are present in MLCK, where they function to localize MLCK to actin filaments. To test the role of the DFRXXL motif in the Myo3A tail, we altered the codon for arginine (R) within the DFRXXL motif to one encoding alanine (A), resulting in a DNA sequence encoding DFAXXL. Cells expressing GFP-Myo3A containing this R-to-A mutation (GFP:M3 R/A; Figure 3C) lacked the fluorescence at filopodial tips seen in cells expressing GFP:M3 (Figure 3B), indicating an intact DFRXXL motif is required for filopodia tip localization. Unlike cells expressing GFP:3THDI,3THDII (Figure 3D), cells expressing a similar tail fragment construct containing the R-to-A mutation (GFP:3THDI,3THDII R/A; Figure 3E) did not exhibit colocalization with actin filaments, indicating an intact DFRXXL motif is required for actin association.
Figure 3.
Arginine-to-Alanine (R/A) point mutation in DFRXXL motif abolishes filopodial localization by full-length myosin IIIA and actin association by the tail domain. (A) Diagram shows location and amino acid sequence of the 3THDII found at the very C-terminus of myosin IIIA proteins. The human and bass 3THDII sequences are shown for comparison. The putative actin-binding motif DFRXXL is boxed in red. The arginine (R) to alanine (A) site-directed mutation within the fish DFRXXL motif is shown. HeLa cells were transfected with GFP constructs and the composite images of GFP fluorescence (green) and F-actin staining (red) are shown: (B) GFP:M3 localizes to the cytoplasm and filopodial tips; (C) GFP:M3 R/A localizes to the cytoplasm but not in filopodial tips; (D) GFP:3THDI,3THDII localizes throughout the cell and colocalizes with actin filaments in the cell body and cell protrusions; (E) GFP:3THDI,3THDII R/A localizes throughout the cell but not with actin structures. Arrowheads point to representative filopodial tips; arrows point to examples of F-actin structures that show (D) or lack (E) colocalization with GFP; bars, 10 μm.
3THDII Interacts with Actin In Vitro
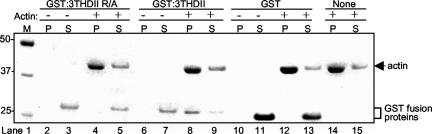
The presence of a DFRXXL motif suggests that the observed association between 3THDII and actin filaments in HeLa cells is direct. To test this idea, we expressed glutathione-S-transferase (GST) fusion proteins containing either wild-type or mutated 3THDII sequences in bacteria and tested the purified fusion proteins for actin binding in vitro using an actin spin-down assay. Fusion proteins were incubated alone or with actin filaments assembled in vitro from purified actin monomers, followed by centrifugation to pellet the actin fibers and visualized by SDS-PAGE. As shown in the Coomassie blue–stained gel in Figure 4 (lanes 6–9), GST: 3THDII was mostly present in the pellet (P) fraction when centrifuged in the presence of F-actin (which pellets) but not in its absence, indicating an interaction between GST:THDII and actin filaments. Altered fusion protein (GST:3THDII R/A; lanes 2–5), which contains the R-to-A amino acid change within the DFRXXL sequence, remained in the supernatant fraction (S) after centrifugation in both the presence or absence of F-actin, indicating an intact DFRXXL motif is required for actin binding. GST protein (GST; lanes 10–13) used as a control remained in the supernatant in both the presence and the absence of actin filaments. These observations demonstrate that the 3THDII directly binds actin filaments, and that this is due to the presence of the DFRXXL motif.
Figure 4.
3THDII binds actin filaments in vitro; R-to-A mutation in 3THDII abolishes actin binding. Actin spin-down assay: GST proteins were incubated with (+) or without (–) actin filaments, centrifuged, and equal portions of the resulting supernatant (S) and pellet (P) fractions were analyzed using SDS-PAGE and Coomassie-blue staining. GST: 3THDII R/A, 3THDII containing arginine-to-alanine (R/A) point mutation in the DFRXXL motif, lanes 2–5; GST:3THDII, wild-type 3THDII, lanes 6–9; GST control, lanes 10–13. Lane 1 contains molecular mass markers with sizes in kDa denoted to the left. Lanes 14 and 15 show control experiments that contained actin filaments but lacked GST proteins.
Myo3A Colocalizes with Calmodulin at Filopodial Tips
Striped bass Myo3A contains 9 IQ motifs whose locations extend beyond the predicted neck region and into the tail domain, and Myo3A has been shown to interact with calmodulin in vitro (Dosé et al., 2003). To determine whether Myo3A interacts with calmodulin in HeLa cells, we expressed a GFP-calmodulin fusion construct (GFP: CaM) alone or with a nontagged version of Myo3A. Cells expressing GFP:CaM exhibited a strong, diffuse fluorescence throughout the cytoplasm and nucleus (Figure 5A), with only 2% of transfected cells (n = 200) showing some filopodial tip fluorescence (unpublished data). In cells expressing both GFP:CaM and (untagged) Myo3A, 39% of the GFP:CaM transfected cells (n = 150) exhibited fluorescence at filopodial tips (Figure 5B). As expected, cells expressing untagged Myo3A alone showed staining of filopodial tips, as determined by immunocytochemical analysis using a Myo3A antibody (Figure 5C). Thus, this Myo3A-dependent localization of calmodulin indicates that Myo3A is bound to one or more calmodulin molecules in HeLa cells.
Figure 5.
Calmodulin localizes to filopodia tips when coexpressed with Myo3A. HeLa cells were transfected with plasmid(s) encoding (A) GFP:CaM alone; (B) GFP:CaM and (untagged) Myo3A; (C) Myo3A alone. (A and B) A composite of GFP fluorescence (green) and F-actin staining (red); (C) a composite of immunodetection (green) of Myo3A by anti-Myo3A antibody and F-actin staining (red). Arrowheads point to representative filopodial tips. Bars, 10 μm.
DISCUSSION
In fish photoreceptor cells, Myo3A concentrates at the distal ends of long, ellipsoidal actin bundles that terminate in the calycal processes (Dosé et al., 2003). Calycal processes are actin-filled, cellular protrusions of unknown function that extend from the inner segment of both rod and cone photoreceptors to form a cup around the base of the outer segment (Nagle et al., 1986; Arikawa and Williams, 1991). NINAC p174, the long-form of the Drosophila class III myosin, concentrates in rhabdomeres, microvillus-like structures that are the site of action of many of the steps in phototransduction in fly photoreceptor cells (Porter et al., 1992). In this report, we have shown that a significant portion of GFP-Myo3A localizes to the distal ends of actin bundles within filopodia in transfected HeLa cells (Figure 2B). Filopodia are thin, cellular protrusions involved in cell motility and are thought to function as sensors of the local environment and as sites for adhesion and signaling (Lewis and Bridgman, 1992; Davenport et al., 1993). Thus, Myo3A localizes to actin filaments present within cellular protrusions in both photoreceptors and transfected HeLa cells.
A mutation in the motor active site (Shimada et al., 1997) of Myo3A was created to test the role of motor activity in myosin III localization. This active-site mutation is predicted to abolish ATP-cleaving activity, thus creating a motor immobilized in an ATP-bound, low actin-affinity, intermediate step of the motor cycle. The motor-altered Myo3A did not localize to filopodia tips (Figure 2C), which suggests Myo3A moves out toward the filopodial tip on the actin bundle under its own power. Similarly, a NINAC protein lacking the complete motor domain failed to localize to the rhabdomeres, which resulted in both ERG and retinal degeneration phenotypes (Porter and Montell, 1993). Because myosin motor activity is unidirectional and the actin bundles in both filopodia and rhabdomeres (Arikawa et al., 1990) are oriented with their plus-ends toward the tips of the processes, these observations suggest that class III myosins are plus-end directed motors. In agreement with this observation, Komaba et al. (2003) recently have reported that a truncated form of human myosin IIIA has plus-end directed, F-actin translocating activity in vitro.
The presence of a kinase domain in class III myosins implies a role in cellular signaling or regulation of motor activity. A mutation in the NINAC kinase domain resulted in normal localization but an altered electroretinogram (ERG) phenotype (Porter and Montell, 1993). In our studies, a truncated Myo3A lacking the kinase domain did not disrupt filopodia tip localization (Figure 2D). These observations suggest that kinase activity is not required for either motor activity or localization. Indeed, the kinase deleted-Myo3A localized more effectively to filopodia tips than the full-length construct, as seen by the elongated fluorescent pattern in the tips and reduced cytoplasmic fluorescence. This suggests that the presence of the kinase domain may inhibit Myo3A filopodia localization to some degree. Filopodia are presumably a major site of action for signal transduction cascades, because filopodial extension, retraction, and adhesion are likely to be highly regulated. The same may be true for the filopodia-like calycal processes, because they have been shown in fish photoreceptors to be dynamic cellular extensions that lengthen and shorten in response to changes in light condition (Pagh-Roehl et al., 1992). Perhaps one of Myo3A's functions is to localize its kinase domain to calycal processes where it participates in some local signal-transduction cascade.
The tail domain of NINAC p174 is required for rhabdomeres localization and for binding to INAD, a PDZ-domain protein that functions as a scaffold for the “signalplex” in Drosophila photoreceptors (Wes et al., 1999). NINAC and myosin IIIA tail domains share no detectable sequence similarity, and the Myo3A tail does not contain a PDZ-binding consensus sequence (Harris and Lim, 2001). We have shown here that the 3THDII, which is highly conserved in vertebrate myosin IIIA tails, contains an actin-binding motif, DFRXXL. The DFRXXL motif was first identified as a novel actin-binding motif present in the N-terminal domain of smooth muscle MLCK (Smith et al., 1999). The role of this motif has not been characterized in any other protein so far. However, Smith et al. (1999) have noted that putative DFRXXL motifs are present in chicken smooth muscle isoform of α-actinin, in Ca2+/calmodulin-dependent protein kinase I, and in titin. The actual role of actin binding by the myosin IIIA tail domain is unclear. An intact DFRXXL sequence is required for Myo3A tail:actin interactions both in HeLa cells (Figure 2E) and in vitro (Figure 4) and for localization of full-length Myo3A to filopodia tips (Figure 3C). This latter observation could be explained by a loss of Myo3A motility or an inability to target to the filopodial tip. The loss of filopodial localization by GPF:M3-inactive, which has an intact tail but altered motor, suggests the change in localization is due to a lack of motility.
The tail domain of class I myosins associates with actin filaments (Lynch et al., 1986). The three class I myosins from Acanthamoeba and two of the five class I myosins from Dictyostelium contain a Gly/Pro/Ala-rich domain, termed Tail Homology 2 (TH2), that binds actin filaments in vitro (Brzeska et al., 1988; Doberstein and Pollard, 1992; Jung and Hammer, 1994). It is thought that tail:actin interactions could serve to recruit type I myosins onto actin filaments to aid processivity. Alternatively, a second actin-binding site could allow the type I myosins to cross-link and contract actin filaments.
It is curious that full-length myosin IIIA does not appear to interact with actin filaments other than those found bundled in cellular protrusions, despite having two actin binding sites, one in the motor and one in the tail. It is known that tropomyosin restricts the interaction of most myosins with stress fibers. However, the Myo3A tail actin-binding site is not blocked from binding stress fibers when expressed as GFP:3THDI or GFP:3THDI,3THDII, (Figure 2, F and G), suggesting that the tail binds to a different site on the actin filament than do myosin motor domains. In fact, Stull and coworkers presented NMR-derived structural evidence that the DFRXXL motifs in smooth muscle MLCK bind to an actin subunit at a unique site, opposite the side where tropomyosin and myosin II bind (Hatch et al., 2001). Perhaps in the context of the full-length Myo3A protein, actin binding by the tail domain is regulated. Indeed, a constitutively active actin-binding motif in the tail might be expected to be detrimental to translocation of a myosin. On the other hand, perhaps weak actin filament interactions mediated by the tail may support translocation by keeping myosin IIIA in close proximity to actin filaments, while the motor domain cycles on and off the actin filament during its power strokes. Such a mechanism may be advantageous for single-headed motors, which is the predicted structure of class III myosins, as they cannot move in a hand-overhand mechanism hypothesized for two-headed motors. Alternatively, the tail actin-binding site may function to limit myosin IIIA motor activity to actin bundles. The tail may interact with adjoining filaments present in actin bundles to promote efficient motor activity. Therefore, without the tail:actin interactions available in bundles, Myo3A would lack sufficient actin interactions for motility. This model would explain the apparent absence of Myo3A on the nonbundled F-actin mesh present at the cell cortex in HeLa cells and nonbundled F-actin in photoreceptors (Dosé and Burnside, 2002).
Three different human MYO3A mutations have been identified that lead to nonsyndromic hearing loss (Walsh et al., 2002). Two of the mutations truncate the MYO3A protein before the tail domain, and a third alters a splice acceptor that leads to an unstable message. Given that NINAC is required in the fly eye and that our results indicate an important role for the tail domain in Myo3A localization, it is surprising that these mutations in human MYO3A do not result in vision problems as well. This suggests Myo3A function is critical in the ear but not in the eye, that Myo3B can compensate for Myo3A function in the retina, or that different splice variants of Myo3A may be critical in the eye and the ear.
Recently, myosin X has been shown to localize to filopodial tips in HeLa cells, where time-lapse imaging revealed it undergoes both forward and rearward movements (Berg and Cheney, 2002). Unlike Myo3A, the filopodial tip localization of myosin X did not require its tail domain. Overexpression of full-length myosin X (but not truncated forms of myosin X) in COS7 cells caused an increase in the number and length of filopodia, indicating that myosin X or its cargo may function in filopodial dynamics. We have not observed any similar changes in filopodia of either HeLa or Cos7 cells transfected with Myo3A constructs.
Given that both myosin X and myosin IIIA localize to the tips of cell protrusions, it is intriguing to speculate that myosin IIIA may function to translocate cargo out to the distal end of the calycal processes. The Myo3A cargo is unknown, but the presence of IQ motifs beyond the neck domain in myosin IIIA proteins suggests calmodulin may be one important cargo. We have demonstrated here that myosin IIIA can concentrate calmodulin in the filopodial tips in HeLa cells (Figure 5B). Calmodulin is a highly conserved regulatory protein that mediates a variety of calcium iondependent signaling pathways. In Drosophila, calmodulin localization in the rhabdomeres is dependent on NINAC myosins, because mutant flies lacking the NINAC p174 did not concentrate calmodulin in the rhabdomere, and this correlated with a defect in vision (Porter et al., 1993). Further work toward identifying and characterizing myosin IIIA cargo is imperative in order to understand the role of Myo3A in vertebrate sensory organs.
Acknowledgments
We thank Ann Fisher and members of the David Drubin and Karsten Weis laboratories in Berkeley for supplying cell lines and giving helpful advice on tissue culture. We thank Jennifer Lin-Jones and other members of the Burnside group for helpful comments throughout this project. This work was supported by National Institutes of Health grant EY-03575.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E02–10–0656. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E02-10-0656.
Abbreviations used: 3THDI, myosin III tail homology domain I; 3THDII, myosin III tail homology domain II; GFP, green fluorescent protein; F-actin, filamentous actin.
References
- Arikawa, K., Hicks, J.L., and Williams, D.S. (1990). Identification of actin filaments in the rhabdomeral microvilli of Drosophila photoreceptors. J. Cell Biol. 110, 1993–1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arikawa, K., and Williams, D.S. (1991). Alpha-actinin and actin in the outer retina: a double immunoelectron microscopic study. Cell Motil. Cytoskel. 18, 15–25. [DOI] [PubMed] [Google Scholar]
- Berg, J.S., and Cheney, R.E. (2002). Myosin-X is an unconventional myosin that undergoes intrafilopodial motility. Nat. Cell Biol. 4, 246–250. [DOI] [PubMed] [Google Scholar]
- Brzeska, H., Lynch, T.J., and Korn, E.D. (1988). Localization of the actin-binding sites of Acanthamoeba myosin IB and effect of limited proteolysis on it actin-activated Mg2+-ATPase activity. J. Biol. Chem. 5, 427–435. [PubMed] [Google Scholar]
- Dan, I., Watanabe, N.M., and Kusumi, A. (2001). The Ste20 group kinases as regulators of MAP kinase cascades. Trends Cell Biol. 11, 220–230. [DOI] [PubMed] [Google Scholar]
- Davenport, R.W., Dou, P., Rehder, V., and Kater, S.B. (1993). A sensory role for neuronal growth cone filopodia. Nature 361, 721–724. [DOI] [PubMed] [Google Scholar]
- Doberstein, S.K., and Pollard, T.D. (1992). Localization and specificity of the phospholipid and actin binding sites on the tail of Acanthamoeba myosin IC. J. Cell Biol. 117, 1241–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dosé, A.C., and Burnside, B. (2000). Cloning and chromosomal localization of a human class III myosin. Genomics 67, 333–342. [DOI] [PubMed] [Google Scholar]
- Dosé, A.C., and Burnside, B. (2002). A class III myosin expressed in the retina is a candidate for Bardet-Biedl syndrome. Genomics 79, 621–624. [DOI] [PubMed] [Google Scholar]
- Dosé, A.C., Hillman, D.W., Wong, C., Sohlberg, L., Lin-Jones, J., and Burnside, B. (2003). Myo3A, one of two class III myosin genes expressed in vertebrate retina, is localized to the calycal processes of rod and cone photoreceptors and is expressed in the sacculus. Mol. Biol. Cell 14, 1058–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goode, B.L., Wong, J.J., Butty, A., Peter, M., McCormack, A.L., Yates, J.R., Drubin, D.G., and Barnes, G. (1999). Coronin promotes the rapid assembly and cross-linking of actin filaments and may link the actin and microtubule cytoskeletons in yeast. J. Cell Biol. 144, 83–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris, B.Z., and Lim, W.A. (2001). Mechanism and role of PDZ domains in signaling complex assembly. J. Cell Sci. 114, 3219–3231. [DOI] [PubMed] [Google Scholar]
- Hatch, V., Zhi, G., Smith, L., Stull, J.T., Craig, R., and Lehman, W. (2001). Myosin light chain kinase binding to a unique site on F-actin revealed by three-dimensional image reconstruction. J. Cell Biol. 154, 611–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodge, T., and Cope, M.J.T.V. (2000). A myosin family tree. J. Cell Sci. 113, 3353–3354. [DOI] [PubMed] [Google Scholar]
- Jung, G., and Hammer, J.A. (1994). The actin binding site in the tail domain of Dictyostelium myosin IC (myoC) resides within the glycine- and proline-rich sequence (tail homology region 2). FEBS Lett. 342, 197–202. [DOI] [PubMed] [Google Scholar]
- Komaba, S., Inoue, A., Maruta, S., Hosoya, H., and Ikebe, M. (2003). Determination of human myosin III as a motor protein having a protein kinase activity. J. Biol. Chem. 278, 21352–21360. [DOI] [PubMed] [Google Scholar]
- Lewis, A.K., and Bridgman, P.C. (1992). Nerve growth cone lamellipodia contain two populations of actin filaments that differ in organization and polarity. J. Cell Biol. 119, 1219–1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch, T.J., Albanesi, J.P., Korn, E.D., Robinson, E.A., Bowers, B., and Fujisaki, H. (1986). ATPase activities and actin-binding properties of subfragments of Acanthamoeba myosin IA. J. Biol. Chem. 261, 17156–17162. [PubMed] [Google Scholar]
- Montell, C., and Rubin, G.M. (1988). The Drosophila ninaC locus encodes two photoreceptor cell specific proteins with domains homologous to protein kinases and the myosin heavy chain head. Cell 52, 757–772. [DOI] [PubMed] [Google Scholar]
- Nagle, B.W., Okamoto, C., Taggart, B., and Burnside, B. (1986). The teleost cone cytoskeleton. Localization of actin, microtubules, and intermediate filaments. Invest. Ophthalmol. Vis. Sci. 27, 689–701. [PubMed] [Google Scholar]
- Ng, K.P., Kambara, T., Matsuura, M., Burke, M., and Ikebe, M. (1996). Identification of myosin III as a protein kinase. Biochemistry 35, 9392–9399. [DOI] [PubMed] [Google Scholar]
- Pagh-Roehl, K., Wang, E., and Burnside, B. (1992). Shortening of the calycal process actin cytoskeleton is correlated with myoid elongation in teleost rods. Exp. Eye Res. 55, 735–46. [DOI] [PubMed] [Google Scholar]
- Porter, J.A., Hicks, J.L., Williams, D.S., and Montell, C. (1992). Differential localizations of and requirements for the two Drosophila ninaC kinase/myosins in photoreceptor cells. J. Cell Biol. 116, 683–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter, J.A., and Montell, C. (1993). Distinct roles of the Drosophila ninaC kinase and myosin domains revealed by systematic mutagenesis. J. Cell Biol. 122, 601–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter, J.A., Yu, M., Doberstein, S.K., Pollard, D., and Montell, C. (1993). Dependence of calmodulin localization in the retina on the NINAC unconventional myosin. Science 262, 103. [DOI] [PubMed] [Google Scholar]
- Shimada, T., Sasaki, N., Ohkura, R., and Sutoh, K. (1997). Alanine scanning mutagenesis of the Switch I region in the ATPase site of Dictyostelium discoideum myosin II. Biochemistry 36, 14037–14043. [DOI] [PubMed] [Google Scholar]
- Smith, L., Su, X., Lin, P.-J., Zhi, G., and Stull, J.T. (1999). Identification of a novel actin binding motif in smooth muscle myosin light chain kinase. J. Biol. Chem. 274, 29433–29438. [DOI] [PubMed] [Google Scholar]
- Walsh, T., Walsh, V., Vreugde, S., Hertzano, R., Shahin, H., Haika, S., Lee, M.K., Kanaan, M., King, M.-C., and Avraham, K.B. (2002). From flies' eyes to our ears: mutations in a human class III myosin cause progressive nonsyndromic hearing loss DFNB30. Proc. Natl. Acad. Sci. USA 99, 7518–7523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wes, P.D., Xu, X.Z., Li, H.S., Chien, F., Doberstein, S.K., and Montell, C. (1999). Termination of phototransduction requires binding of the NINAC myosin III and the PDZ protein INAD. Nat. Neurosci. 2, 447–453. [DOI] [PubMed] [Google Scholar]