Abstract
Gp210 is an evolutionarily conserved membrane protein of the nuclear pore complex (NPC). We studied the phenotypes produced by RNAi-induced downregulation of gp210 in both human (HeLa) cells and Caenorhabditis elegans embryos. HeLa cell viability requires Gp210 activity. The dying cells accumulated clustered NPCs and aberrant membrane structures at the nuclear envelope, suggesting that gp210 is required directly or indirectly for nuclear pore formation and dilation as well as the anchoring or structural integrity of mature NPCs. Essential roles for gp210 were confirmed in C. elegans, where RNAi-induced reduction of gp210 caused embryonic lethality. The nuclear envelopes of embryos with reduced gp210 also had aberrant nuclear membrane structures and clustered NPCs, confirming that gp210 plays critical roles at the nuclear membrane through mechanisms that are conserved from nematodes to humans.
INTRODUCTION
In all eukaryotes, two nuclear membranes separate the chromosomes from the cytoplasm. Communication depends on nuclear pore complexes (NPCs), which are positioned at sites of fusion between the inner and outer membranes. NPCs are protein complexes with an estimated mass of 60 MDa in yeast and 125 MDa in vertebrates (reviewed in Pante and Aebi, 1996; Vasu and Forbes, 2001). Each NPC consists of multiple copies of nearly 30 distinct proteins (Rout et al., 2000; Cronshaw et al., 2002) termed nucleoporins, which collectively maintain pore structure and mediate molecular transport between nucleoplasm and cytoplasm (reviewed in Gorlich and Kutay, 1999; Macara, 2001; Lei and Silver, 2002).
Of the ∼30 distinct nucleoporins identified in a comprehensive proteomic analysis of vertebrate NPCs, only two (gp210 and Pom121) are integral membrane proteins (Cronshaw et al., 2002). Of these two, only gp210 is detectable in both animals and plants, suggesting that its function has been conserved in evolution (Cohen et al., 2001). Very little of gp210 is exposed to the cytoplasm. There is a short (∼60-residue) C-terminal tail, which is exposed and faces the NPC; gp210 then crosses the membrane once, and most of its mass (∼1750 N-terminal residues) is positioned in the nuclear envelope lumen (Greber et al., 1990). Gp210 forms stable dimers, and possibly higher-order oligomers (Favreau et al., 2001). The lumenal domain of rat gp210 includes an amphiphilic helix, which is potentially similar to “fusion” peptides of viral proteins that stimulate membrane fusion (Greber et al., 1990) and its C-terminal domain has a left-handed polyproline type II (PII) helical structure, which is implicated in protein-protein interactions (Pilpel et al., 2003). Several previous studies have addressed the ultrastructural location of gp210 in mature NPCs, its targeting to nuclear pore membranes, and cell cycle dynamics (reviewed by Bodoor et al., 1999). The hypothesis that gp210 might be required to form nuclear pores was recently tested using cell-free extracts of Xenopus eggs (Drummond and Wilson, 2002). These experiments focused on the function of the exposed tail of gp210, which was accessible to reagents. This study showed that the addition of competing gp210 tail polypeptides blocked the growth of nascent nuclei, due to dominant inhibition of pore formation. In these arrested nuclei, apparently fused but undilated “mini-pores” accumulated at the nuclear envelope. Antibodies against the exposed gp210 tail also inhibited pore formation; however in this case, the arrested intermediates were too electron-dense to determine if the membranes had actually fused. Under both conditions, the arrested nuclei also accumulated “twinned” membrane structures, characterized by short regions of close apposition of the inner and outer membranes (Drummond and Wilson, 2002). These results suggested that the gp210 tail mediates the dilation of nascent pores and hinted that gp210 might also directly mediate “porogenic” membrane fusion. These experiments did not address the function of the lumenal domain of gp210.
To understand the function of gp210, we determined its downregulation phenotypes in both human cultured cells and Caenorhabditis elegans embryos. This strategy eliminates the entire gp210 protein in vivo, rather than dominantly inhibiting its exposed domain in vitro. We report here that gp210 is an essential protein in both human and C. elegans cells, where it appears to be required directly or indirectly for pore formation and the stability of mature NPCs.
MATERIALS AND METHODS
Strains, Antibodies, and Materials
C. elegans Bristol N2 animals were maintained under standard conditions as described (Brenner, 1974). Rabbit polyclonal antibodies against the rod and tail domains of Ce-lamin and rabbit polyclonal antibodies against human emerin were described previously (Liu et al., 2000; Lee et al., 2001). To generate polyclonal antibodies against Ce-Gp210, rats were immunized at 3-week intervals with either of two antigens. Rat 3783 was immunized with a synthetic peptide conjugated to keyhole limpet hemocyanin (KLH). This peptide, NH2-CDKTGSFGDNTLNNTTH-COOH (Gp210-C peptide), comprised Ce-Gp210 residues 1777–1792 plus an N-terminal cysteine. Peptide synthesis, purification by reverse-phase HPLC with the use of a C18 analytical column, and conjugation to KLH were done by Boston Biomolecules (Woburn, MA). Rat 3586 was immunized with a gel-purified recombinant polypeptide corresponding to exon 3 of Ce-gp210 fused to GST. Immunizations and serum production were done by Covance Research Products (Denver, PA). mAb 414, which recognizes five FG-repeat nucleoporins (nup358, nup214/CAN, nup153, nup98, and p62), was purchased from BABCO (Richmond, CA). Anti-human gp210 antibodies were kindly provided by Dr. J.C. Courvalin (Courvalin et al., 1990). Rabbit polyclonal antibodies against human lamins A/C were kindly provided by Dr. K. Weber (Harborth et al., 2001). Polyclonal antibodies specific for lamin B2 were kindly provided by Dr. G. Krohne (Wurzburg). All secondary antibodies were purchased from Jackson Laboratories (West Grove, PA). Epon was purchased from Agar Scientific (Stansted, Essex, UK).
dsRNA-mediated Interference Templates in C. elegans and HeLa Cells
Full-length Ce-lamin cDNA (Liu et al., 2000) and DNA fragments corresponding to Ce-gp210 exons 3–5 (including introns) or Ce-gp210 exons 8–15 (without introns; kindly provided by Dr. Y. Kohara, Japan) were subcloned into feeding vector L4440 (Timmons and Fire, 1998) and used in the RNAi feeding experiments. The Ce-gp210 fragments were also subcloned into pBlueScript KS and used as templates to prepare dsRNA for gp210(RNAi) soaking experiments. The dsRNA oligonucleotides used to inhibit the human gp210 gene (5′-AAUGCGUGCGAAUCCTCACUdTdT-3′) corresponded to nucleotides 523–543 of the human gp210 coding sequence (accession number: BC020573). These oligos were designed as described (Elbashir et al., 2002) and purchased from Dharmacon (Lafayette, CO) in deprotected and desalted form. As a control we used a nonspecific duplex of firefly luciferase from Photinus pyralis (GL2) at the same final concentrations as the gp210 RNA duplex.
RNA-mediated Inhibition (RNAi) in C. elegans
Feeding experiments were done using an optimal protocol (Kamath et al., 2001), with 1 mM isopropyl-beta-d-thiogalactoside. Residual protein in gp210(RNAi) embryos was detected by indirect immunofluorescence as described (Liu et al., 2000). For soaking experiments, single-strand RNA was transcribed in vitro using RiboMAX (Promega, Madison WI), according to the manufacturer protocol. RNA was phenol-extracted twice, ethanol-precipitated, and resuspended in soaking buffer (10.9 mM Na2HPO4, 5.5 mM KH2PO4, 2.1 mM NaCl, 4.7 mM NH4Cl, 3 mM spermidine, and 0.05% gelatin). The two complementary strands of RNA were annealed by incubating 15 min at 68°C, followed by 30 min at 37°C, before transferring worms into the solution. Soaking was done as described (Maeda et al., 2001).
Cell Culture and siRNA Transfection
Cultured HeLa cells were transfected with siRNA as described (Elbashir et al., 2002) with a few minor changes. In short, human HeLa SS6 cells were grown at 37°C in DMEM supplemented with 10% FCS, penicillin, and streptomycin. Cells were passed regularly to maintain exponential growth. The day before transfection, cells were trypsinized, diluted with fresh medium without antibiotics, and transferred to 24-well plates (532 μl per well). Transient transfection of siRNAs was done using TransIT-TKO Transfection Reagent (Mirus Corporation, Madison, WI). For each well, we first preincubated 50 μl OPTIMEM 1 medium (Gibco, Grant Island, NY) plus 3 μl TransIT-TKO for 5 min at room temperature. Separately, 12 μl OPTIMEM 1 medium was mixed with 3 μl siRNA duplex. The two mixtures were combined and incubated 15 min at room temperature to allow complex formation for a final concentration of 100 nM siRNA.
Indirect Immunofluorescence
Unless otherwise stated, all steps were done at room temperature. Staining of C. elegans by indirect immunofluorescence was described previously (Gruenbaum et al., 2002). Transfected HeLa SS6 cells grown on glass coverslips in 24-well plates were fixed in methanol 6 min at –20°C, washed with PBS, and blocked in 10% milk in PBS for 1 h. Cells were incubated 1 h with gp210 antibodies (Courvalin et al., 1990) diluted 1:100 in PBS containing 10% milk (PBSM). Slides were then washed three times (10 min each) in PBS, incubated 1 h with Cy3-conjugated secondary antibodies diluted 1:200 in PBS, and washed in PBS three times (10 min each). Slides were then incubated 1 h with antibodies against lamin A/C, lamin B2, emerin, or FG-repeat nucleoporins diluted 1:200, 1:200, 1:100, and 1:50, respectively, in PBSM. Slides were washed as described and incubated 1 h with the appropriate FITC-conjugated secondary antibody diluted 1:200 in PBS, washed once in PBS for 10 min, incubated 10 min with 10 μg/ml DAPI, and washed once in PBS. Slides were then mounted in glycerol containing 2% N-propyl-gallate.
SDS-PAGE and Immunoblotting
Mixed stage N2 animals were recovered from plates by washing in 1 ml PBS, transferred to a 1.5-ml Eppendorf tube, mixed with 1 ml 2× sample loading buffer (25 mM Tris-HCl, pH 6.8, 20% glycerol, 0.2 M β-mercaptoethanol, 4% SDS, 0.001% bromophenol blue), frozen at –70°C for at least 30 min and then boiled for 5 min before loading on SDS-PAGE. Proteins were separated on 5–17% SDS-PAGE gradient gels and transferred to nitrocellulose membranes in buffer lacking SDS to facilitate the transfer of high-molecular-weight proteins. Nonspecific binding was blocked in PBST containing 5% nonfat dry milk overnight and probed with antibodies against Ce-Gp210 (sera 3783 or 3586, diluted 1:500), in PBST-milk for 2 h at 22–24°C. After washing three times in PBST, blots were incubated 1 h at 22–24°C with horseradish peroxidase–conjugated secondary antibodies. Blots were washed as described above and visualized by enhanced chemiluminescence (Amersham Pharmacia Biotech).
Sample Preparation for TEM Analysis
C. elegans embryo samples were prepared exactly as described (Cohen et al., 2002). HeLa cells were grown in a sterile chamber slide with cover (Nunc, Naperville, IL) for 3 d after the siRNA transfection and fixed in 2.5% glutaraldehyde plus 3% formaldehyde in 0.1 M sodium cacodylate (pH 7.4) for 1 h at room temperature. Samples were washed four times (10 min each) in 0.1 M sodium cacodylate (pH 7.4) and then incubated 1 h at room temperature in osmium tetroxide solution (1% OsO4, 1.5% K3Fe(CN)6, 0.1 M sodium cacodylate, pH 7.4). Samples were then washed four times (10 min each) in sodium cacodylate pH 7.4, and then dehydrated by sequential incubation in 30%, 50%, 70%, 90%, and 95% ethanol (10 min each), followed by three washes (30 min each) in 100% ethanol. These dehydrated samples were then (a) incubated 2 h at room temperature in 25% Epon in ethanol, (b) incubated overnight in 50% Epon in ethanol, (c) incubated in 75% Epon in ethanol for 8 h, (d) incubated overnight in 100% Epon, and finally incubated for 8 h in fresh 100% Epon. Epon block was polymerized for 2 d at 60°C in a dry oven and then sectioned to give 80–90-nm thin sections, using a Diatome diamond knife. Sections were picked up on 200-mesh thin bar copper grids, stained with uranyl acetate and lead citrate, and viewed with an electron microscope (Philips Tecnai 12; www.feic.com/tecnai/advanc.htm) equipped with Mega-View II CCD camera, and analySIS version 3.0 software (Soft Imaging System GmbH, Münster, Germany).
FACS Analysis and Cell Sorting
HeLa cells were transfected with siRNA targeting gp210 and harvested after 5 d. Harvested cells were washed with sterile PBS, fixed in 100% methanol overnight at 4°C, washed in PBS, incubated with 100 μl of 100 μg/ml RNAse and 50 μg/ml propidium iodide (Sigma, Dorset, United Kingdom), incubated on ice for 10 min, and then stored at 4°C until FACS analysis. FACS analysis was performed on a FACSCalibur system (Becton-Dickinson, San Jose, CA). The left edge of the control cell profile was taken as the border separating normal, diploid cells (to the right) from apoptotic, hypodiploid cells (to the left). The percentage of apoptotic cells was calculated as the ratio of events on the left side to events from the whole population as described (Benhar et al., 2001).
Nuclear Import Assays
HeLa SS6 cells were cultivated on coverslips for 3 d after siRNA transfection and then treated with 33 μg/ml digitonin for 4.5 min. As the transport substrate, we assayed BSA conjugated to both rhodamine and a nuclear localization signal peptide (NLS-BSA) as described (Broder et al., 1997). Translocation of fluorescent NLS-BSA conjugates into nuclei of digitonin-permeabilized HeLa cells was assayed by confocal microscopy.
Cell Growth and Survival
Two days after siRNA treatment, cells were harvested, counted and replated in equal numbers in triplicates in 24-well plates, and left to grow as described above. Each day, one of the triplicates was fixed for 10 min at room temperature with 0.5% glutaraldehyde (Merck, Frankfurt, Germany), washed three times, 10 s each, with water, once for 1 min in 0.1 M borate (pH 8.5) and then incubated for 1 h in 1% methylene blue (Sigma). Wells were then washed in running distilled water till washing water ran clear and completely air-dried. The stain was extracted in 0.1 N HCl for 1 h in 37°C. Absorbance was read in an enzyme-linked immunosorbent assay (ELISA) reader at 630 nm as described (Shir and Levitzki, 2002).
RESULTS
Gp210 Is Essential in HeLa Cells
The term “pore” will refer specifically to the membrane structure (hole), not the proteinaceous NPC assembled therein. To determine the downregulation phenotype for gp210 in vivo, we first used the siRNA technique (Elbashir et al., 2002) to reduce gp210 expression in human tissue culture (HeLa) cells. Cells were transfected with a 21-base pair dsRNA corresponding to nucleotides 523–543 in the human gp210 coding sequence, and 99.7% (n = 1564) of these “gp210-siRNA” cells had a severely reduced gp210 immunofluorescence signal (Figure 1A). Control cells transfected with dsRNA corresponding to the luciferase gene (GL2-siRNA cells) had normal immunofluorescence signals for gp210 (Figure 1A). Survival assays showed that gp210 siRNA treatment reduced cell proliferation and was lethal to most cells within 5 d (Figure 1B). Likewise, propidium iodide staining and FACS analysis performed 5 d after transfection showed that more than 60% of the gp210 siRNA-treated cells were dead, compared with 6% of untreated cells (unpublished data). The dying cells were in sub-G1 suggesting apoptosis, while the remaining cells were divided normally between the G1 and G2 phases. We then analyzed the cultures by double staining with gp210 antibodies and trypan blue dye, which stains dead cells. This analysis showed that 91% (n = 96) of trypan blue–positive cells in the gp210-siRNA culture were negative for gp210, and the remaining 9% had significantly reduced levels of gp210 staining. In contrast, all trypan blue–positive cells in the GL2-siRNA culture had normal levels of gp210 (unpublished data). We therefore concluded that gp210 was essential for the viability of dividing cells.
Figure 1.
Phenotypes of HeLa cells with reduced gp210. (A) Transfection of HeLa cells with dsRNA specific for human gp210 gene (gp210-siRNA) caused significantly reduced expression of gp210 relative to control cells transfected with nonspecific dsRNA (GL2-siRNA). Cells were double-stained for gp210 (Courvalin et al., 1990) by indirect immunofluorescence and DAPI to visualize DNA and imaged by fluorescence microscopy. (B) The growth rates of gp210-siRNA and untreated HeLa cells were quantified by a colorimetric assay (see MATERIALS AND METHODS). The graph shows the ratio of OD(630) measurements for gp210-siRNA cells vs. untreated cells at different time points. The exponential regression is monotonously descending (with a negative exponential coefficient) showing that the growth rate of gp210-siRNA cells was lower than that of untreated cells at all times. (C) Downregulation of gp210 disrupts FG-repeat nucleoporins. Gp210-siRNA cells and control GL2-siRNA cells were double-stained for human gp210 and FG-repeat nucleoporins (mAb414) and visualized by laser confocal microscopy. The exposure times shown for immunostaining are identical within each set of panels in A and C. (D) Thin-section transmission electron micrographs of GL2-siRNA control cells (a and b) and gp210-siRNA cells (c–f), 3 d after siRNA treatment. Cells with reduced gp210 had condensed chromatin (c), clustering of mature NPCs (d), enlarged nuclear envelope lumenal space (e), and clusters of pore-related membrane structures (f). Arrows in D depict mature NPCs (b and d), membrane blebs (e), and various arrested pore structures (f), which were named “twinned membranes” (t) and “fusion-arrested” membranes (f), as described in text. Bars: 30 μm in A, 20 μm in C, 200 nm in D (b, d–f), and 2000 nm in D (a and c).
Gp210 Is Required for the Assembly or Maintenance (or Both) of Nuclear Pore Complexes
Indirect immunofluorescence staining showed that nuclear envelopes with reduced levels of gp210 also stained weakly for FG repeat nucleoporins, detected using mAb 414 (mAb414; Figure 1C). These results suggested that loss of gp210 might have either direct or downstream effects on the assembly, recruitment or stability of FG-repeat nucleoporins, which are positioned throughout the NPC.
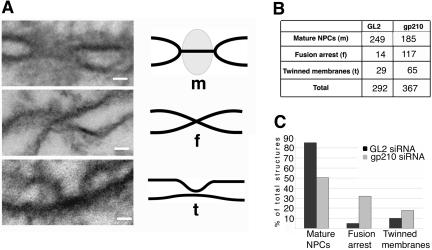
To visualize nuclear envelope morphology in gp210-siRNA cells, we used thin-section transmission electron microscopy (TEM). Other cells from the same samples were verified as gp210-deficient by immunofluorescence staining, using gp210 antibodies (unpublished data). GL2-siRNA cells served as positive controls. Five days after siRNA transfection most cells had died, exhibiting typical apoptotic morphology including condensed chromatin, blebbed membranes, and disrupted mitochondrial structure (unpublished data). We therefore analyzed gp210-siRNA cell cultures 3 d after transfection, when cell growth and viability were reduced but many cells were still alive (Figure 1B) and attached to culture wells. The control (GL2-siRNA) nuclei had typical nuclear envelopes and widely spaced mature NPC structures (Figure 1D, a and b). In contrast, the gp210-siRNA nuclei had reproducibly aberrant phenotypes including abnormally condensed and abnormally distributed chromatin (Figure 1Dc) and clustered NPCs; 43% of analyzed nuclei (n = 81) had clusters in which three or more NPCs were located within 250 nm of their nearest neighbor (Figure 1Dd), compared with 0% (n = 38) of control GL2-siRNA cells (Figure 1Db). Additional phenotypes included the enlargement of the lumenal space between the inner and outer membranes (Figure 1De) and the accumulation of aberrant pore-related membrane structures (Figures 1Df and 2A). These aberrant structures, termed “twinned membranes” and “fusion-arrested,” were similar to those caused by dominant inhibition of the gp210 tail (Drummond and Wilson, 2002), so we adapted similar nomenclature. The arrested membrane structures are shown at higher magnification in Figure 2A. Twinned membranes consisted of short (∼40 nm) regions where the inner and outer membranes were unusually close (Figure 2A, t). Most fusion-arrested membranes formed an × structure for which we could not determine if membrane fusion had occurred (Figure 2A, f), but a minor subset had rounded membrane profiles connected by a dense “bridge” ∼25-nm diameter (unpublished data). We quantified these structures and mature NPCs in TEM micrographs of gp210-siRNA and control GL2-siRNA cells. Compared with controls, the gp210-siRNA cells had fewer mature NPCs (50 vs. 85%) and significantly more fusion-arrested structures (32 vs. 5%) and twinned membranes (18 vs. 10%). The change in the distribution of mature NPCs and pore-related structures was highly significant (chi-square test; p « 0.00001). It was interesting that 50% of the NPCs still appeared normal 3 d after gp210-siRNA treatment. We hypothesize that as cells become depleted of gp210, cooperative interactions between remaining gp210 proteins might allow a subset of NPCs to survive. We speculate that the aberrant nuclear membrane structures, comprising 50% of total pore-related structures, might represent 1) mature NPCs that degenerated because of insufficient gp210 subunits and 2) arrested intermediates in the formation of new pores.
Figure 2.
Quantitation of pore-related nuclear membrane structures in gp210-siRNA HeLa cells. We identified (A) and quantified (B and C) three distinct morphological features including mature NPCs (m), fusion-arrested structures (f), and twinned membranes (t) on TEM micrographs of gp210-siRNA cells and control GL2-siRNA HeLa cells. The number of counted structures for each category is shown in B, and graphed in C as the percentage of total structures counted in either gp210-siRNA or control GL2-siRNA cells. Bars in A, 50 nm.
Three days after treatment, the gp210-siRNA cells had a significantly lower immunofluorescence signal for lamins A/C at the nuclear envelope, but a similar or higher lamin A/C signal in the nucleoplasm (Figure 3Ab). The nuclear envelope signal for lamin B2 was slightly reduced, compared with matched GL2-siRNA control cells (Figure 3Aa). No difference was detected for emerin, an integral membrane protein that localizes by diffusion within the plane of the ER/nuclear membrane and is subsequently retained at the inner nuclear membrane by binding to stable structures (Holmer and Worman, 2001; Figure 3Ac). These results hinted that gp210 downregulation might affect either the efficiency of lamin import into nuclei or lamin assembly. We therefore tested for nuclear import in the gp210-siRNA cells. Three days after transfection, gp210- and GL2-siRNA cells were permeabilized with digitonin and incubated with a fluorescence import-competent substrate (rhodamine-nuclear localization signal peptide-BSA, referred to as NLS-BSA) and then fixed and analyzed by confocal microscopy. All cells remaining attached to the coverslips were considered live cells. In control GL2-siRNA cells, the NLS-BSA substrate was efficiently imported into nuclei, as seen by the enriched rhodamine signal at the nuclear envelope and inside the nucleus (Figure 3B, middle). Other controls showed no nuclear signal in unpermeabilized GL2-siRNA cells, as expected (Figure 3B, right), or when import was assayed at 4°C or blocked by treatment with wheat germ agglutinin (unpublished data). We then examined import in gp210-siRNA cells, and were somewhat surprised by the answer. The fluorescent substrate was found predominantly in the cytoplasm and nuclear envelope, with a weak signal in the nucleoplasm (Figure 3B, left). The fluorescent signal was strongest at the nuclear envelope, suggesting that the fluorescent substrate could bind NPCs, but failed to translocate efficiently. This result suggested that reduced levels of gp210 could also compromise the import activity of mature-appearing NPCs.
Figure 3.
Localization of nuclear lamins and emerin in gp210-siRNA cells. (A) Immunofluorescence of GL2-siRNA and gp210-siRNA cells double-stained for gp210 plus either lamin B (a), lamin A/C (b), or emerin (c). Images were obtained by laser confocal microscopy. (B) Assay for in vitro import of rhodamine-labeled NLS-BSA into nuclei of digitonin-permeabilized gp210-siRNA cells (left), GL2-siRNA cells (middle), or unpermeabilized cells (right). Each arrow indicates one nucleus. Bars: 20 μm in A and 10 μm in B.
C. elegans gp210 Is Associated with NPCs
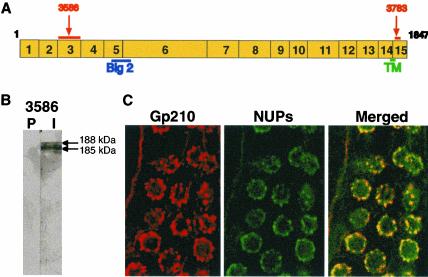
To determine if the essential roles of gp210 have been conserved in evolution, we studied gp210 in C. elegans (Ce-Gp210). We previously identified a gene encoding Ce-gp210 on C. elegans chromosome I, at position 1.02 (Cohen et al., 2001). This gene, previously named npp-12 in WormBase (http://www.wormbase.org) encodes a predicted protein of 1847 residues with a predicted mass of 209 kDa and one predicted membrane-spanning region near its C termini (Figure 4A), similar to vertebrate gp210. Ce-Gp210 has nine potential sites for N-linked glycosylation, plus a predicted bacterial Ig-like Type 2 (Big2) domain. Interestingly, Big2 domains are characteristic of Escherichia coli intimins and other bacterial and bacteriophage surface proteins (Kelly et al., 1999). To our knowledge gp210 is the only eukaryotic protein that shares this domain (Cohen et al., 2001).
Figure 4.
Gp210 in C. elegans. (A) A schematic diagram of full-length Ce-Gp210 (residues 1–1847), drawn approximately to scale, showing the exons (numbered boxes), a single predicted transmembrane domain (TM), and the bacterial Ig-like type 2 (Big2) domain. Regions used as antigen are marked by orange lines and labeled by serum number (3586 and 3783). (B) Western blot of crude protein extract of mixed stage C. elegans probed with preimmune (P) or immune (I) serum 3586. (C) Confocal images of an adult C. elegans gonad double-stained by indirect immunofluorescence using Ce-Gp210 serum 3783 (left panel), FG-repeat nucleoporins (mAb414; NUPs; middle panel), and the merged images (right panel).
We raised rat polyclonal antibodies against a C-terminal peptide of Ce-gp210 (serum 3783) and a recombinant polypeptide comprising exon 3 (residues 102–223) of Ce-Gp210 (serum 3586; see MATERIALS AND METHODS). On immunoblots of C. elegans total protein lysates, both sera specifically recognized a major band that migrated at 185 kDa plus a minor band at 188 kDa (Figure 4B shows results for serum 3586). We do not know why two bands are present, but speculate that differential glycosylation, alternative splicing, or proteolysis could produce them. Recognition of both proteins was specifically competed by pretreating antibodies with the peptide antigen (unpublished data). The antipeptide serum 3783 gave stronger signals in immunofluorescence assays and was used in the following indirect immunofluorescence experiments. Ce-Gp210 localized at the nuclear rim in C. elegans gonads in a punctate pattern characteristic of NPCs, and colocalized with the family of FG-repeat nucleoporins recognized by mAb mAb414 (Davis and Blobel, 1986; Figure 4C). C. elegans fed with the lmn-1(RNAi) construct produced lamin-depleted embryos in which the Ce-gp210 signal became clustered (unpublished data), consistent with its effect on FG-repeat nucleoporins (Liu et al., 2000). We concluded that our antibodies specifically recognized endogenous Ce-Gp210 proteins at the NPC and could be used to analyze the down-regulation phenotype of Gp210 in C. elegans.
Gp210 Is Essential for Viability in C. elegans Embryos, and Its Loss Produces Aberrant Nuclear Membrane Structures and NPC Spacing Phenotypes
In two previous studies, wild-type C. elegans were either fed or soaked in dsRNA corresponding to various exons of Ce-gp210 (Fraser et al., 2000; Maeda et al., 2001). Fraser et al. (2000) fed C. elegans with dsRNA corresponding to exons 5–9 and found no phenotypes. In contrast, Maeda et al. (2001) used a dsRNA soaking strategy that targeted exons 8–15 of Ce-gp210; this approach was lethal to 36% of the embryos and caused all remaining animals to arrest as larvae. However, both studies lacked controls for specific loss of Ce-Gp210 protein. To determine definitively the RNAi-induced downregulation phenotype for Gp210, we fed, injected, or soaked nematodes with dsRNA corresponding to different combinations of Ce-Gp210 exons. By indirect immunofluorescence, Ce-Gp210 protein was reduced only in F1 embryos that were treated with dsRNA that included exons 3–5 (unpublished data) or exons 8–15 (Figure 5A). Feeding and injection methods were both less efficient than soaking, which produced 15% embryonic lethality before the comma stage. The remaining embryos hatched and developed normally and were fertile and indistinguishable from wild-type animals. All arrested gp210(RNAi) embryos had little or no detectable Ce-Gp210, whereas animals that escaped embryonic lethality stained positive for Ce-Gp210 (unpublished data). We concluded that the RNAi method was not effective for all embryos. Nevertheless, the early lethality associated with reduced Ce-Gp210 protein showed conclusively that Ce-gp210 is essential in C. elegans.
Figure 5.
RNAi-induced downregulation phenotype for Ce-Gp210. (A) Double immunostaining of control embryos (left) or Cegp210(RNAi) embryos (right; exons 8–15 were targeted), imaged by fluorescence microscopy. Embryos were stained by indirect immunofluorescence using antibodies against Ce-Gp210 (serum 3783), Ce-lamin (Liu et al., 2000), or FG-repeat nucleoporins (Nups; mAb414). Arrows indicate NPC clusters. (B) Thin-section TEMs of control untreated embryos (a) and Ce-gp210(RNAi) embryos in which exons 3–5 were targeted (b and c). Cegp210(RNAi) embryos had more condensed chromatin (b), an enlarged nuclear envelope lumen (b), and clustered NPCs (c). NU, nucleus; Cyt, cytoplasm. Bars: 10 μm (A), 200 nm (B, c), 750 nm (B, b), and 1500 nm (B, a).
To determine if other nucleoporins were affected by loss of Ce-Gp210, Ce-gp210(RNAi) embryos were double-stained by indirect immunofluorescence using antibodies against endogenous Ce-Gp210 plus either endogenous FG-repeat nucleoporins or Ce-lamin. Ce-lamin localized normally in the absence of Ce-Gp210 (Figure 5A, gp210(RNAi)). Thus, the subtle effect on lamin B localization seen in HeLa cells was not detectable in C. elegans. The main difference between the C. elegans and human cell phenotypes was that FG-repeat nucleoporins remained relatively abundant at the nuclear envelopes of gp210-reduced C. elegans cells. However, these nucleoporins were partially clustered as seen by indirect immunofluorescence staining (Figure 5A, arrows in gp210(RNAi)) and at higher resolution by TEM (Figure 5Bc). Thus, NPC clustering was a consistent feature of gp210-depletion in both worm and human cells (Figure 1D). These results collectively suggested that Gp210 is directly or indirectly required for the spacing, anchoring, and structural stability of NPCs.
To analyze nuclear ultrastructure in C. elegans cells lacking Ce-Gp210, we collected F1 embryos that were produced after feeding with dsRNA against Ce-Gp210 (targeting exons 3–5). These F1 embryos were fixed and visualized by thin-section TEM (see MATERIALS AND METHODS). Control embryos were obtained from animals fed with “empty” L4440 vector. Nuclei in the Ce-gp210(RNAi) embryos showed several aberrant phenotypes including abnormally condensed chromatin (Figure 5Bb), NPC clustering (Figure 5Bc) and subtle enlargement of the nuclear envelope lumen (Figure 5Bb). These phenotypes were not seen in control embryos (Figure 5Ba). The lethality and associated nuclear and NPC phenotypes seen in gp210-depleted C. elegans embryos were all consistent with the phenotypes of gp210-depleted HeLa cells, strongly suggesting that gp210 functions have been conserved in evolution.
DISCUSSION
The results of this study show that gp210 is essential for the viability of dividing cells. Gp210 was required for nuclear pore assembly and dilation as well as for anchoring of mature NPCs. The downregulation phenotypes for gp210 were very similar in C. elegans embryos and human cell cultures, suggesting that gp210 function has been conserved in evolution. The amino acid sequences of gp210 in C. elegans and mammals (rat) are only 25% identical (44% similar) (Cohen et al., 2001). These identical residues are likely to be essential for the structure or function of gp210 and will serve as starting points for future molecular analysis.
Gp210 Is Involved in NPC Spacing
In both human cells and C. elegans, nuclei with reduced gp210 failed to space NPCs evenly around the nuclear envelope, as evidenced by NPC clustering. Given the large volume of the nucleus, an even distribution of NPCs is probably important for efficient nuclear import and export. The mechanisms of NPC spacing are not understood. Nuclear lamins are critical for spacing (Lenz-Bohme et al., 1997; Liu et al., 2000; Schirmer et al., 2001), but how NPCs attach to lamin filaments is unclear. One possibility is suggested by the interaction between Nup153 and lamins and a demonstrated role for Nup153 in NPC anchoring (Smythe et al., 2000; Walther et al., 2001). We cannot determine from our experiments if gp210 is directly involved in anchoring NPCs. However, given that its mass is primarily lumenal, we suggest that the loss of gp210 disrupts other nucleoporins with direct roles in NPC anchoring. Indeed, NPC clustering phenotypes are caused by mutations in many yeast nucleoporins (Zabel et al., 1996; Bucci and Wente, 1998; Siniossoglou et al., 2000; Ryan and Wente, 2002), some of which have vertebrate homologues (reviewed in Vasu and Forbes, 2001).
Similarities in the gp210 Downregulation Phenotypes in Human and C. elegans Cells
Knockdown of the gp210 gene in both human HeLa cells and C. elegans embryos caused nuclear membranes to accumulate aberrant structures termed twinned and fusion-arrested membranes. These aberrant structures could represent a mixture of arrested intermediates in porogenic nuclear membrane fusion and degenerating mature NPCs. There were many similarities between the C. elegans and human cell phenotypes, including cell death, chromatin disruptions, the accumulation of aberrant nuclear membrane structures and NPC clustering. These similarities collectively suggest that the loss of gp210 cause both direct and downstream effects, due to loss of NPCs.
Aberrant Membrane Structures in gp210-depleted Cells: Parallels with HA-mediated Fusion
Our findings could not distinguish between degenerating preexisting NPCs and arrested intermediates. However, the formation of abnormal membrane structures could support models in which gp210 is involved in nuclear membrane fusion events. It is therefore interesting to consider recent studies with HA (hemagglutinin), the fusogenic protein encoded by influenza virus, and other fusogenic membrane proteins (Skehel and Wiley, 2000). Kozlov and Chernomordik (2002) have proposed a model that may account for why so many (hundreds to thousands) of molecules of HA are required for efficient membrane fusion and for pore dilation to biologically significant diameters. In this model, large numbers of HA molecules aggregate laterally to form a “coat” that cooperatively undergoes a conformational change into a curved surface, which pushes membranes together and provides the driving force for membrane fusion and dilation. Fusion can still occur with fewer HA molecules, but the resulting products are arrested in either of two states. One state is “hemi-fused”: here, only the inner leaflets fuse, forming a membrane “bridge” that blocks content mixing between the two compartments. A second arrested state involves complete fusion of both leaflets, but the resulting pore fails to dilate (Kozlov and Chernomordik, 2002).
Our results for cells that are running out of gp210 are consistent with such models. We speculate that twinned membranes might represent the earliest detectable stage of arrest, with the fewest gp210 molecules, because these intermediates had pronounced curvature of one membrane (almost always the outer membrane) in close contact with the second membrane, but lacked a “focal” point for fusion. We propose that fusion-arrested pores have enough gp210 to initiate, but not enough to complete, the fusion reaction. Note that twinned and fusion-arrested membrane structures were also produced in assembling nuclei, by treatment with competing gp210 tail polypeptides and antibodies directed against the Xenopus gp210 tail domain, respectively (Drummond and Wilson, 2002). Overall, our results strongly support a direct role for gp210 in nuclear membrane fusion and are consistent with current models in which multiple copies of the fusogenic protein (e.g., gp210) are required to efficiently fuse two flattened membranes and dilate the resulting pore. Interestingly, our results suggest additional roles for gp210 in maintaining the spacing and structural integrity of mature NPCs. Further studies to test these models will shed light on a fusion mechanism fundamental to all nucleocytoplasmic communication in eukaryotes.
Acknowledgments
We thank J.C. Courvalin for the anti-human gp210 antibodies and K. Weber and G. Krohne for antibodies against human lamin. We also thank A. Loyter for his help in performing the import assay. This work was funded by grants from the Israel Science Foundation (ISF) and Austrian Bank (to Y.G.), the National Institutes of Health (GM64535 to K.L.W.), and a Journal of Cell Science travel award and Boeringer Ingelheim travel allowance (to M.C.).
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E03–04–0260. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E03-04-0260.
References
- Benhar, M., Dalyot, I., Engelberg, D., and Levitzki, A. (2001). Enhanced ROS production in oncogenically transformed cells potentiates c-Jun N-terminal kinase and p38 mitogen-activated protein kinase activation and sensitization to genotoxic stress. Mol. Cell. Biol. 21, 6913–6926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodoor, K., Shaikh, S., Enarson, P., Chowdhury, S., Salina, D., Raharjo, W.H., and Burke, B. (1999). Function and assembly of nuclear pore complex proteins. Biochem. Cell Biol. 77, 321–329. [PubMed] [Google Scholar]
- Broder, Y.C., Stanhill, A., Zakai, N., Friedler, A., Gilon, C., and Loyter, A. (1997). Translocation of NLS-BSA conjugates into nuclei of permeabilized mammalian cells can be supported by protoplast extract. An experimental system for studying plant cytosolic factors involved in nuclear import. FEBS Lett. 412, 535–539. [DOI] [PubMed] [Google Scholar]
- Brenner, S. (1974). The genetics of Caenorhabditis elegans. Genetics 77, 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucci, M., and Wente, S.R. (1998). A novel fluorescence-based genetic strategy identifies mutants of Saccharomyces cerevisiae defective for nuclear pore complex assembly. Mol. Biol. Cell 9, 2439–2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen, M., Tzur, Y.B., Neufeld, E., Feinstein, N., Delannoy, M.R., Wilson, K.L., and Gruenbaum, Y. (2002). Transmission electron microscope studies of the nuclear envelope in C. elegans embryos. J. Struct. Biol. 140, 232–240. [DOI] [PubMed] [Google Scholar]
- Cohen, M., Wilson, K.L., and Gruenbaum, Y. (2001). Membrane proteins of the nuclear pore complex: Gp210 is conserved in vertebrates, Drosophila melanogaster, C. elegans and Arabidopsis. Palo Alto, CA: Gene Teraphy Press.
- Courvalin, J.C., Lassoued, K., Bartnik, E., Blobel, G., and Wozniak, R.W. (1990). The 210-kD nuclear envelope polypeptide recognized by human autoantibodies in primary biliary cirrhosis is the major glycoprotein of the nuclear pore. J. Clin. Invest. 86, 279–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronshaw, J.M., Krutchinsky, A.N., Zhang, W., Chait, B.T., and Matunis, M.J. (2002). Proteomic analysis of the mammalian nuclear pore complex. J. Cell Biol. 158, 915–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis, L.I., and Blobel, G. (1986). Identification and characterization of a nuclear pore complex protein. Cell 45, 699–709. [DOI] [PubMed] [Google Scholar]
- Drummond, S.P., and Wilson, K.L. (2002). Interference with the cytoplasmic tail of gp210 disrupts “close apposition” of nuclear membranes and blocks nuclear pore dilation. J. Cell Biol. 158, 53–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbashir, S.M., Harborth, J., Weber, K., and Tuschl, T. (2002). Analysis of gene function in somatic mammalian cells using small interfering RNAs. Methods 26, 199–213. [DOI] [PubMed] [Google Scholar]
- Favreau, C., Bastos, R., Cartaud, J., Courvalin, J.C., and Mustonen, P. (2001). Biochemical characterization of nuclear pore complex protein gp210 oligomers. Eur. J. Biochem. 268, 3883–3889. [DOI] [PubMed] [Google Scholar]
- Fraser, A.G., Kamath, R.S., Zipperlen, P., Martinez-Campos, M., Sohrmann, M., and Ahringer, J. (2000). Functional genomic analysis of C. elegans chromosome I by systematic RNA interference. Nature 408, 325–330. [DOI] [PubMed] [Google Scholar]
- Gorlich, D., and Kutay, U. (1999). Transport between the cell nucleus and the cytoplasm. Annu. Rev. Cell Dev. Biol. 15, 607–660. [DOI] [PubMed] [Google Scholar]
- Greber, U.F., Senior, A., and Gerace, L. (1990). A major glycoprotein of the nuclear pore complex is a membrane-spanning polypeptide with a large lumenal domain and a small cytoplasmic tail. EMBO J. 9, 1495–1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruenbaum, Y., Lee, K.K., Liu, J., Cohen, M., and Wilson, K.L. (2002). The expression, lamin-dependent localization and RNAi depletion phenotype for emerin in C. elegans. J. Cell Sci. 115, 923–929. [DOI] [PubMed] [Google Scholar]
- Harborth, J., Elbashir, S.M., Bechert, K., Tuschl, T., and Weber, K. (2001). Identification of essential genes in cultured mammalian cells using small interfering RNAs. J. Cell Sci. 114, 4557–4565. [DOI] [PubMed] [Google Scholar]
- Holmer, L., and Worman, H.J. (2001). Inner nuclear membrane proteins: functions and targeting. Cell Mol. Life Sci. 12–13, 1741–1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath, R.S., Martinez-Campos, M., Zipperlen, P., Fraser, A.G., and Ahringer, J. (2001). Effectiveness of specific RNA-mediated interference through ingested double-stranded RNA in Caenorhabditis elegans. Genome Biol. 2, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly, G., Prasannan, S., Daniell, S., Fleming, K., Frankel, G., Dougan, G., Connerton, I., and Matthews, S. (1999). Structure of the cell-adhesion fragment of intimin from enteropathogenic Escherichia coli. Nat. Struct. Biol. 6, 313–318. [DOI] [PubMed] [Google Scholar]
- Kozlov, M.M., and Chernomordik, L.V. (2002). The protein coat in membrane fusion: lessons from fission. Traffic 3, 256–267. [DOI] [PubMed] [Google Scholar]
- Lee, K.K., Haraguchi, T., Lee, R.S., Koujin, T., Hiraoka, Y., and Wilson, K.L. (2001). Distinct functional domains in emerin bind lamin A and DNA-bridging protein BAF. J. Cell Sci. 114, 4567–4573. [DOI] [PubMed] [Google Scholar]
- Lei, E.P., and Silver, P.A. (2002). Protein and RNA export from the nucleus. Dev. Cell 2, 261–272. [DOI] [PubMed] [Google Scholar]
- Lenz-Bohme, B., Wismar, J., Fuchs, S., Reifegerste, R., Buchner, E., Betz, H., and Schmitt, B. (1997). Insertional mutation of the Drosophila nuclear lamin dm(0) gene results in defective nuclear envelopes, clustering of nuclear pore complexes, and accumulation of annulate lamellae. J. Cell Biol. 137, 1001–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, J., Rolef-Ben Shahar, T., Riemer, D., Spann, P., Treinin, M., Weber, K., Fire, A., and Gruenbaum, Y. (2000). Essential roles for Caenorhabditis elegans lamin gene in nuclear organization, cell cycle progression, and spatial organization of nuclear pore complexes. Mol. Biol. Cell 11, 3937–3947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macara, I.G. (2001). Transport into and out of the nucleus. Microbiol. Mol. Biol. Rev. 65, 570–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda, I., Kohara, Y., Yamamoto, M., and Sugimoto, A. (2001). Large-scale analysis of gene function in Caenorhabditis elegans by high-throughput RNAi. Curr. Biol. 6, 171–176. [DOI] [PubMed] [Google Scholar]
- Pante, N., and Aebi, U. (1996). Molecular dissection of the nuclear pore complex. Crit. Rev. Biochem. Mol. Biol. 31, 153–199. [DOI] [PubMed] [Google Scholar]
- Pilpel, Y., Bogin, O., Brumfeld, V., and Reich, Z. (2003). Polyproline type II conformation in the C-terminal domain of the nuclear pore complex protein gp210. Biochemistry 42, 3519–3526. [DOI] [PubMed] [Google Scholar]
- Rout, M.P., Aitchison, J.D., Suprapto, A., Hjertaas, K., Zhao, Y., and Chait, B.T. (2000). The yeast nuclear pore complex: composition, architecture, and transport mechanism. J. Cell Biol. 148, 635–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan, K.J., and Wente, S.R. (2002). Isolation and characterization of new Saccharomyces cerevisiae mutants perturbed in nuclear pore complex assembly. BMC Genet. 3, 17. Available at www.biomedcentral.com/1471-2156/3/17. [DOI] [PMC free article] [PubMed]
- Schirmer, E.C., Guan, T., and Gerace, L. (2001). Involvement of the lamin rod domain in heterotypic lamin interactions important for nuclear organization. J. Cell Biol. 153, 479–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shir, A., and Levitzki, A. (2002). Inhibition of glioma growth by tumor-specific activation of double-stranded RNA-dependent protein kinase PKR. Nat. Biotechnol. 20, 895–900. [DOI] [PubMed] [Google Scholar]
- Siniossoglou, S., Lutzmann, M., Santos-Rosa, H., Leonard, K., Mueller, S., Aebi, U., and Hurt, E. (2000). Structure and assembly of the Nup84p complex. J. Cell Biol. 149, 41–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skehel, J.J., and Wiley, D.C. (2000). Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin. Annu. Rev. Biochem. 69, 531–569. [DOI] [PubMed] [Google Scholar]
- Smythe, C., Jenkins, H.E., and Hutchison, C.J. (2000). Incorporation of the nuclear pore basket protein Nup153 into nuclear pore structures is dependent upon lamina assembly: evidence from cell-free extracts of Xenopus eggs. EMBO J. 19, 3918–3931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmons, L., and Fire, A. (1998). Specific interference by ingested dsRNA. Nature 395, 854. [DOI] [PubMed] [Google Scholar]
- Vasu, S.K., and Forbes, D.J. (2001). Nuclear pores and nuclear assembly. Curr. Opin. Cell Biol. 13, 363–375. [DOI] [PubMed] [Google Scholar]
- Walther, T.C., Fornerod, M., Pickersgill, H., Goldberg, M., Allen, T.D., and Mattaj, I.W. (2001). The nucleoporin Nup153 is required for nuclear pore basket formation, nuclear pore complex anchoring and import of a subset of nuclear proteins. EMBO J. 20, 5703–5714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zabel, U., Doye, V., Tekotte, H., Wepf, R., Grandi, P., and Hurt, E.C. (1996). Nic96p is required for nuclear pore formation and functionally interacts with a novel nucleoporin, Nup188p. J. Cell Biol. 133, 1141–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]