Abstract
We show that Cdc6, an essential initiation factor for DNA replication, undergoes caspase-3–mediated cleavage in the early stages of apoptosis in HeLa cells and SK-HEP-1 cells induced by etoposide, paclitaxel, ginsenoside Rh2, or tumor necrosis factor-related apoptosis-inducing ligand. The cleavage occurs at the SEVD442/G motif and generates an N-terminal truncated Cdc6 fragment (p49-tCdc6) that lacks the carboxy-terminal nuclear export sequence. Cdc6 is known to be phosphorylated by cyclin A-cyclin dependent kinase 2 (Cdk2), an event that promotes its exit from the nucleus and probably blocks it from initiating inappropriate DNA replication. In contrast, p49-tCdc6 translocation to the cytoplasm is markedly reduced under the up-regulated conditions of Cdk2 activity, which is possibly due to the loss of nuclear export sequence. Thus, truncation of Cdc6 results in an increased nuclear retention of p49-tCdc6 that could act as a dominant negative inhibitor of DNA replication and its accumulation in the nucleus could promote apoptosis. Supporting this is that the ectopic expression of p49-tCdc6 not only promotes apoptosis of etoposide-induced HeLa cells but also induces apoptosis in untreated cells. Thus, the caspase-mediated cleavage of Cdc6 creates a truncated Cdc6 fragment that is retained in the nucleus and induces apoptosis.
INTRODUCTION
The Cdc6 protein plays an essential role in the initiation of DNA replication at discrete chromosomal locations during the S phase of the cell cycle (Dalton and Whitbread, 1995; Lei et al., 1996; Williams et al., 1997; Yan et al., 1998). Overexpression of Cdc6 induces endoreplication and hence polyploidization in HEL cells, Schizosacchromyces pombe, and Arabidopsis thaliana (Nishitani and Nurse, 1995; Castellano et al., 2001; Bermejo et al., 2002). In S. pombe, overexpression of Cdc6/Cdc18 results in rereplication without mitosis (Nishitani and Nurse, 1995), whereas depletion of Cdc6/Cdc18 prevents replication initiation and results in reductional mitosis in which cells randomly segregate their unreplicated chromosomes (Piatti et al., 1995). Cdc6 is phosphorylated by cyclin A-cyclin dependent kinase 2 (cyclin A-Cdk2) at N-terminal CDK phosphorylation sites (Jiang et al., 1999; Petersen et al., 1999), an event that is thought to facilitate its translocation from the nucleus to the cytoplasm and hence prevent reinitiation of replication during the S and G2 phases (Petersen et al., 1999; Delmolino et al., 2001). The conformational changes induced by Cdc6 phosphorylation may unmask the C-terminal nuclear export signal (NES) required for the nuclear export (Delmolino et al., 2001). Overexpression of a mutant form of Cdc6/Cdc18 lacking CDK phosphorylation sites leads to the nuclear accumulation of the protein and a dramatic increase in rereplication (Liang and Stillman, 1997). Thus, rereplication of chromosomes can be induced in the absence of cell division by a mechanism that involves preventing the cytoplasmic translocation of Cdc6.
The rereplication of chromosomes seems to be functionally linked to apoptotic cell death because polyploid cells undergo apoptosis after prolonged spindle disruption (Casenghi et al., 1999; Hong et al., 1999; Verdoodt et al., 1999). This suggests that aberrant Cdc6 regulation may contribute to apoptosis induction. A growing body of evidence suggests that an important feature of apoptosis is the disruption of the link between DNA synthesis and CDK activity that results in the unscheduled activation of CDKs in the presence of unreplicated or partially replicated chromosomes (Shi et al., 1994; reviewed in Guo and Hay, 1999; Hakem et al., 1999; Jin et al., 2000). Recent studies have demonstrated that CDKs, which are cell cycle regulators, play an essential role in executing the apoptosis in human umbilical vein endothelial cells, HeLa, and SK-HEP-1 cells that is induced by various apoptosis-inducing reagents (reviewed in King and Cidlowski, 1998; Park et al., 1998; Hakem et al., 1999; Levkau et al., 1999; Jin et al., 2000). It may be that the up-regulation of CDK activity during apoptosis causes an unscheduled phosphorylation of Cdc6, which may subsequently induce an unscheduled cytoplasmic translocation of phospho-Cdc6 in the cells. Intriguingly, during the early stages of apoptosis induced by tumor necrosis factor-α and cycloheximide, human Cdc6 is rapidly cleaved by the caspase-dependent pathway; cleavage is also ensured by a separate proteasome-dependent pathway in cells undergoing apoptosis induced by the DNA-damaging drug adozelesin (Blanchard et al., 2002). The cleavage of Cdc6 has been suggested to be a primordial response that uncouples DNA replication from the cell cycle (Blanchard et al., 2002). However, the means by which Cdc6 is cleaved and the functional relevance of this event to apoptosis are not fully understood. Thus, we asked whether the subcellular localization of Cdc6 protein is affected by the up-regulation of cyclin A–Cdk2 activity or by the proteolytic cleavage during apoptosis and if so, whether these alterations might be functionally linked to the induction of apoptosis.
Herein, we show that Cdc6 undergoes site-specific cleavage by a caspase-3-like protease during the early stages of apoptosis in HeLa cells and SK-HEP-1 cells treated with several inducers of apoptosis. We found that this cleavage resulted in the nuclear accumulation of a Cdc6 fragment that contains all known sites necessary for forming prereplication complex. That ectopic expression of the cleaved fragment (p49-tCdc6) not only markedly promotes apoptosis in etoposide-treated cells but also induces apoptosis in untreated cells suggests that the accumulation of p49-tCdc6 contributes to the induction of apoptosis.
MATERIALS AND METHODS
Materials
Ginsenoside Rh2 (G-Rh2) was kindly provided by Dr. D.H. Kim (Kyung Hee University, Seoul, Korea). Etoposide and paclitaxel were obtained from Sigma-Aldrich (St. Louis, MO), whereas necrosis factor-related apoptosis-inducing ligand (TRAIL) and the caspase-3 inhibitor z-DEVD-fmk were from Calbiochem (San Diego, CA). Other caspase inhibitors, the fluorogenic caspase substrate, and recombinant caspases were from BD Biosciences (San Jose, CA). Other chemical reagents were from Sigma-Aldrich. DMEM and calf serum were from Invitrogen (Carlsbad, CA). The transfection reagent Polyfect was from QIAGEN (Valenica, CA) and annexin-V-Alexa 568 was from Roche Diagnostics (Indianapolis, IN).
Cell Culture and Induction of Apoptosis
Human cervical carcinoma (HeLa) and human hepatoma (SK-HEP-1) cells were maintained at 37°C and 5% CO2 as a monolayer culture in DMEM supplemented with 10% (vol/vol) heat-inactivated calf serum, 100 U ml–1 penicillin, 100 μg ml–1 streptomycin, and 250 ng ml–1 amphotericin B. Cells (1 × 106) were seeded on 100-mm Petri dishes and incubated for 24 h at 37°C. The medium was then exchanged with 10% calf serum-supplemented DMEM containing 85 μM etoposide, 80 nM paclitaxel, or 43 μM TRAIL. For treatment with G-Rh2, the culture medium was exchanged with serum-free DMEM containing G-Rh2. HeLa cells were treated with z-DEVD-fmk in the presence of 12 μM G-Rh2 for 2 h.
Preparation of Nuclear Extracts and Immunoblot Analysis
Nuclear extracts were prepared from cells in lysis buffer (10 mM HEPES, pH 7.4, 10 mM KCl, 2 mM MgCl2, 5 mM EGTA, 25 μg ml–1 leupeptin, 5 μg ml–1 pepstatin A, 1 mM phenylmethylsulfonyl fluoride [PMSF], 40 mM β-glycerophosphate, 1 mM dithiothreitol [DTT]) as described previously (Buckley et al., 1999). Briefly, nuclei were collected by centrifugation through 30% sucrose (800 × g, 10 min at 4°C) and resuspended in radioimmunoprecipitation assay buffer (150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, 50 mM Tris pH 8.0, 20 mM β-glycerophosphate, 50 mM NaF, 1 μg ml–1 leupeptin, 1 μg ml–1 antipain, 1 μg ml–1 pepstatin A, 0.1 mM PMSF). For Western blot studies, nuclear lysates were centrifuged at 12,000 rpm for 15 min at 4°C, and the supernatants were collected. After adjusting the protein concentration (bicinchoninic acid assay; Pierce Chemical, Chester, United Kingdom), cell lysates were boiled and resolved by 12% SDS-PAGE before Western blot analysis with appropriate antibodies. Anti-Cdc6 monoclonal antibody raised against full-length Cdc6 of human origin (Santa Cruz Biotechnology, Santa Cruz, CA). Polyclonal anti-poly(ADP-ribose) polymerase (PARP), anti-Cdk2, and monoclonal anti-cyclin A were from Santa Cruz Biotechnology. Monoclonal anti-FLAG was from Upstate Biotechnology (Lake Placid, NY). Immune complexes were revealed using ECL Western blotting detection reagents (Amersham Biosciences, Piscataway, NJ).
Plasmids
Human Cdc6 cDNA and the vector pCS2+GFP were kindly provided by Drs. R.S. Williams (University of Texas Southwestern Medical Center, Dallas, TX) and K.Y. Lee (Chungbuk National University, Cheongju, Korea), respectively. For in vitro translation, Cdc6 cDNA was generated by PCR using 5′-AAGGATCCCCTCAAACCCGATCCCAG-3′ and 5′-AAGGATCCTTAAGGCAATCC AG-3′ (with added BamHI sites) and inserted into pCITE-4c (+) (Novagen, Madison, WI). Converting the Asp 502, Asp 442, Asp 404, Asp 324, and Asp 286 residues into Asn was accomplished by altering the Asp codons GAT or GAC into the Asn codons AAT or AAC, respectively, by the megaprimer method (Sambrook and Russell, 2001). The mutation was first introduced by polymerase chain reaction (PCR) with mutagenic primers (5′-GCGGCTGTGAACCAGTCAGAG-3′, 5′-TCAGAA GTTAATGGTAACAGG-3′, and 5′-GTAGAGTCAAATGTCAAAAGC-3′), the reverse primer (5′-AAGGATCCTTAAGGCAATCCAG-3′), and the forward primer (5′-AAGG ATCCCCTCAAACCCGATCCCAG-3′) and the products were cloned into pCITE-4c (+). All mutant sequences were confirmed. To monitor mammalian expression, we fused Cdc6 to green fluorescent protein (GFP) or FLAG by amplifying full-length Cdc6 cDNA (encoding aa 1–560) or Cdc6 1–442 cDNA (encoding aa 1–442) and inserting the products in-frame into pCS2+GFP or pCS2+FLAG. Full-length human cyclin A cDNA generated by reverse transcription-PCR (by using oligo 5′-AAGGATCCAGCCTATTCTTTGGCC-3′ and 5′-AAGGATCCGGCAG CTGGCATCATTAATAC-3′) with BamHI site at the 5′and 3′ ends was cloned into the pCMV expression vector. Human Cdk2-dn cDNA was kindly provided by Dr. Ewen Mark (Dana-Farber Cancer Institute, Harvard Medical School, Boston, MA).
Coupled In Vitro Transcription/Translation of Cdc6 and In Vitro Cleavage Assay
The in vitro transcription and translation of cDNAs encoding the human Cdc6 protein and the Cdc6 substitution mutants were carried out with the TNT-coupled reticulocyte lysate system (Promega, Madison, WI) according to the manufacturer's protocols. For in vitro cleavage by recombinant caspases, 2 μl of [35S]methionine-labeled Cdc6 was incubated with 100 ng of recombinant caspases and 3 μl of 10× assay buffer (1× buffer: 20 mM HEPES pH 7.5, 10 mM KCl, 1.5 mM MgCl2, 1 mM EDTA, 1 mM EGTA, 0.1 mM PMSF) at 30°C for 1 h in the presence or absence of caspase inhibitors. The total reaction volume was 30 μl. Cleavage products were resolved by 12% SDS-PAGE and visualized by autoradiography.
Transfection and Fluorescence Microscopy Analysis
HeLa cells were seeded on 35-mm culture plates or slides at a density of 5 × 104 cells ml–1 for 24 h before transfection with 3 μg of the appropriate plasmids using Polyfect (QIAGEN) according to the manufacturer's instructions. Live cells were visualized by fluorescence microscopy (IMT-2; Olympus, Tokyo, Japan). For 4,6-diamidino-2-phenylindole (DAPI) staining, cells were fixed in 4% paraformaldehyde solution (Sigma-Aldrich, St. Louis, MO) for 10 min at room temperature 36 h after transfection, treated with DAPI solution (Partec, Münster, Germany) for 10 min at 37°C, and washed twice with phosphate-buffered saline (PBS). Images were made with a camera connected to a fluorescence microscope (BH2-RFCA; Olympus).
Fluorescence-activated Cell Sorting (FACS) Analysis
Cells expressing GFP-tagged proteins were treated with or without 85 μM etoposide and then were washed in PBS, trypsinized, fixed in 75% ethanol, stained with 500 μl of 50 μg/ml propidium iodide solution, and subjected to FACS analysis (particle analyzing System PAS-III; Partec). To differentiate the transfected cells from untransfectants, transfected cells were sorted by a technique of gating a subpopulation of cells by forward scatter versus side scatter, followed by gating green fluorescence intensity. GFP-expressed cells were selectively analyzed for the DNA contents with propidium iodide intensity.
Annexin-V Assay
Cells expressing GFP-tagged proteins were treated with or without 85 μM etoposide for 12 h and then were trypsinized, washed with PBS, and divided into two tubes. One sample was incubated with 2 μl of annexin-V-Alexa 568 in 100 μl of incubation buffer (10 mM HEPES/NaOH pH 7.4, 140 mM NaCl, 5 mM CaCl2) for 20 min at room temperature. And, the other sample was resuspended in 100 μl of the incubation buffer containing 100 ng of propidium iodide for 20 min at room temperature. The percentage of annexin-V–positive cells and propidium iodide-negative cells among the GFP-expressed cells was then determined by flow cytometric analysis (particle analyzing system PAS-III; Partec). Selective analysis of transfected cells was based on gating a subpopulation of cells by forward scatter versus side scatter, followed by gating green fluorescence intensity. Because the percentages of annexin-V(+) cells indicate the percentages of total death-cells and those of PI(+) cells indicate the sum of death-cells in the late apoptosis and necrosis cells, the apoptotic index of the early apoptotic cell populations was calculated by subtracting % PI(+) from % annexin-V(+).
Fluorometric Caspase-3 Activity Assay
Transfected cell lysates (50 μg) were incubated with 200 nM Ac-DEVD-AFC in reaction buffer (20 mM HEPES pH 7.4, 100 mM NaCl, 10 mM DTT, 0.1% CHAPS, 10% sucrose) at 37°C for 1 h. The reaction was monitored by fluorescence emission at 535 nm (excitation at 405 nm).
Cotransfection
HeLa cells were transfected in 35-mm dishes with 1.5 μg of the appropriate plasmids by using 12 μl Polyfect (QIAGEN) according to the manufacturer's instructions and harvested 24 h later in lysis buffer (0.5% Triton X-100, 20 mM Tris pH 7.5, 2 mM MgCl2, 1 mM DTT, 1 mM EGTA, 50 mM β-glycerophosphate, 25 mM NaF, 1 mM Na3VO4, 2 μg ml–1 leupeptin, 2 μg ml–1 pepstatin A, 100 μg ml–1 PMSF, 1 μg ml–1 antipain). Translocation analysis of HeLa cells was performed 24 h after transfection by using the DAPI staining solution as described above.
RESULTS
The Cdc6 Protein Is Specifically Cleaved Early in Apoptosis by a Caspase-3-like Protease to Produce a 49-kDa Fragment
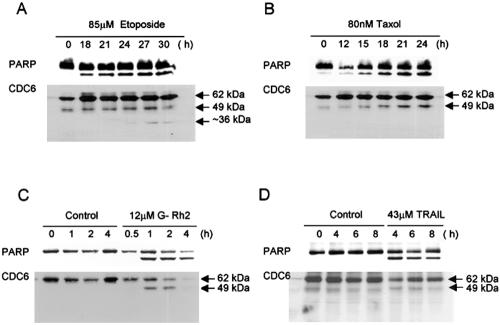
We first monitored the fate of Cdc6 during apoptosis in etoposide-treated HeLa cells. Immunoblot analysis with nuclear extracts showed that the 62-kDa Cdc6 molecule is specifically cleaved into a smaller fragment with a relative molecular weight of 49 kDa with kinetics similar to that of the proteolytic cleavage of PARP (Figure 1A). Similar observations were made with HeLa cells treated with paclitaxel or ginsenoside Rh2 (G-Rh2) and SK-HEP-1 cells treated with TRAIL (Figure 1, B–D). In the etoposide-treated cells, the 49-kDa fragment of Cdc6 was further cleaved into a smaller 36-kDa fragment during later stages of apoptosis, but this cleavage was not detected in the other cases. The Cdc6 cleavage induced by G-Rh2 treatment was prevented by adding the caspase 3 inhibitor, z-DEVD-fmk, to the cell culture (Figure 2A).
Figure 1.
Cdc6 undergoes specific cleavage in HeLa and SK-HEP-1 cells treated with different inducers of apoptosis. HeLa cells or SK-HEP-1 cells were treated for the indicated times. HeLa cells were treated with 85 μM etoposide (A), 80 nM paclitaxel (B), and 12 μM G-Rh2 (C). (D) SK-HEP-1 cells were treated with 43 μM TRAIL. Nuclear extracts were resolved by 12% SDS-PAGE and subjected to immunoblot analysis by using Cdc6- and PARP-specific antibodies.
Figure 2.
In vivo and in vitro caspase-3–dependent cleavage of Cdc6. (A) HeLa cells were treated with 12 μM G-Rh2 in the presence or absence of 100 μM z-DEVD-fmk. Nuclear extracts were subjected to immunoblot analysis as described in Fig. 1. (B) Apoptotic cell extracts were prepared from cells treated with 85 μM etoposide for 24 h (ET), 80 nM paclitaxel for 21 h (TX), or 12 μM G-Rh2 for 2 h (Rh). Cell extracts were incubated at 30°C for 1 h with [35S]methionine-labeled Cdc6 in the presence or absence of 100 μM Ac-DEVD-cho, Ac-VEID-cho, or Ac-IETD-cho. Reaction mixtures were analyzed by 12% SDS-PAGE and visualized by autoradiography. (C) [35S]Methionine-labeled Cdc6 was incubated with recombinant caspases 3, 6, 7, 8, or 9 in the presence or absence of Ac-DEVD-cho at 30°C for 1 h. After resolution by 12% SDS-PAGE, [35S]methionine-labeled Cdc6 was visualized by autoradiography.
We next examined the effect of incubating in vitro-translated [35S]methionine-labeled Cdc6 with extracts of cells treated with etoposide, paclitaxel, or G-Rh2 (Figure 2B). We found that Cdc6 was cleaved by all extracts, and this cleavage was effectively prevented by cotreatment with the inhibitor of caspases 3 and 7 (Ac-DEVD-cho), whereas the caspase 6 inhibitor (Ac-VEID-cho) and the caspase 8 inhibitor (Ac-IETD-cho) had no such effect (Figure 2B). Furthermore, when labeled Cdc6 was incubated with various recombinant caspases, it was efficiently cleaved into fragments with the apparent molecular weights of 49 and 36 kDa by caspases 3 and 7 but not by caspases 6, 8, and 9 (Figure 2C). This cleavage was completely prevented by Ac-DEVD-cho (Figure 2C). Thus, Cdc6 protein is specifically cleaved by a caspase-3-like protease during the early stages of apoptosis.
Identification of the Caspase-3-dependent Cdc6 Cleavage Site
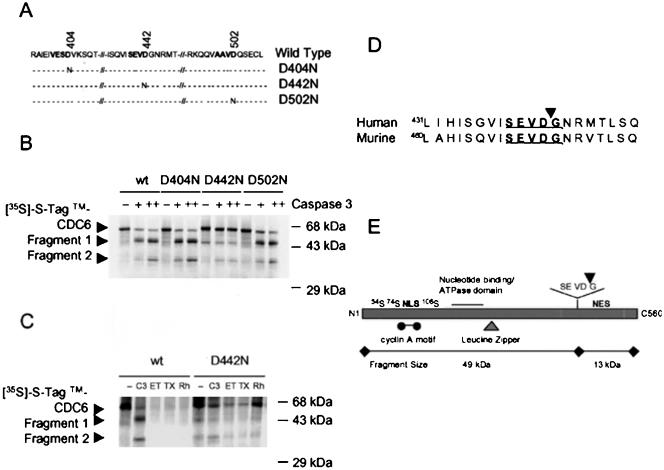
To identify the Cdc6 cleavage site, we searched for a caspase-3 cleavage motif (DXXD↓ followed by a small amino acid) in a region whose cleavage would generate a 49-kDa fragment. We found three possible candidate sequences, namely, VESD404/V, SEVD442/G, and AAVD502/Q (Figure 3A). We used site-directed mutagenesis to substitute asparagine for the aspartic acid residues in each of the candidate sites and generated three [35S]methionine-labeled Cdc6 mutant proteins, D502N, D442N, and D404N, by in vitro translation (Figure 3A). Recombinant caspase-3 efficiently cleaved wild-type Cdc6 (Cdc6-wt) and the D502N and D404N mutant proteins into both fragments, 49 and 36 kDa (Figure 3B). In contrast, the D442N mutant was much less efficiently cleaved, and those cleavages that did occur generated only the 36-kDa fragment. In addition, although Cdc6-wt was degraded by incubation with apoptotic cell lysates induced by etoposide, paclitaxel, or G-Rh2 treatment, the D442N mutant protein was markedly resistant to this degradation (Figure 3C). In an attempt to identify the cleavage site resulting in the 36-kDa fragment, we tested whether DEMD286/Q and DLTD324/R, which have a DXXD motif, can be cleaved by caspase-3. All mutant proteins were cleaved by recombinant caspase-3 as well as Cdc6-wt, indicating that these sites are not caspase-3 cleavage sites (our unpublished results).
Figure 3.
Identification of the caspase-3–dependent Cdc6 cleavage site. (A) Schematic representation of wild-type and mutant Cdc6 proteins. Putative caspase-3 cleavage sites were mutated by using PCR methods; mutant sequences were confirmed. (B) Wild-type and mutant Cdc6 proteins were translated in vitro in the presence of [35S]methionine. Wild-type and mutant [35S]methionine-labeled Cdc6 proteins were incubated at 30°C for 1 h with recombinant caspase-3. The reaction mixtures were analyzed by autoradiography as described in Figure 2. (C) In vitro cleavage assay of [35S]methionine-labeled wild-type and mutant Cdc6 proteins in the presence of recombinant caspase-3 or G-Rh2-, etoposide-, or paclitaxel-treated HeLa cell lysates. The reaction conditions and analysis were as described in Figure 3B. (D) Comparison of Cdc6 amino acid sequences at putative cleavage sites in different species. (E) The caspase-3 cleavage site and functional domains of human Cdc6 responsible for interactions with other proteins (Herbig et al., 1999; Petersen et al., 1999; Takei et al., 1999; Takayama et al., 2000; Delmolino et al., 2001).
Interestingly, the caspase-3 cleavage site SEVD442/G is located near an NES motif in the Cdc6 carboxy terminus and is also found in the murine Cdc6 protein (Figure 3D). Moreover, the N-terminal p49-tCdc6 fragment that results from cleavage at this site contains most of the known Cdc6 functional domains (Herbig et al., 1999; Petersen et al., 1999; Takei et al., 1999; Takayama et al., 2000; Delmolino et al., 2001), including the cyclin A-Cdk2 phosphorylation sites, the NLS motif, the ATPase domain, and the leucine zipper (Figure 3E). This fragment lacks only the carboxy-terminal NES motif (Figure 3E). The pattern of these structural features led us to hypothesize that p49-tCdc6 can enter the nucleus and form a prereplication complex but cannot be exported to the cytoplasm during apoptosis.
The p49-tCdc6 Is Efficiently Phosphorylated by Cyclin A-Cdk2 in HeLa Cells but Its Cytoplasmic Translocation Is Markedly Suppressed
We next tested whether translocation of the p49-tCdc6 protein to the cytoplasm is perturbed due to the absence of the NES, causing it to be trapped in the nucleus. To this end, HeLa cells were transfected with pCS2+ plasmids expressing GFP, GFP-tagged Cdc6-wt, or p49-tCdc6 protein together with pCMV plasmids expressing mock, cyclin A, or Cdk2-dn. The subcellular locations of the GFP-tagged proteins were then assessed (Figure 4, A and B). In the mock-transfected cells, Cdc6-wt was localized in the nucleus in 70.5% of the cells and in the cytoplasm in 29.5% of the cells. However, when cyclin A was coexpressed, these subcellular localizations were markedly shifted to 5.7% in the nucleus and 94.3% in the cytoplasm. In contrast, when Cdk2-dn, a dominant negative mutant version of Cdk2, was coexpressed with Cdc6-wt, the latter protein was in the nucleus in 99.4% of the cells and in the cytoplasm in only 0.6% of the cells (Figure 4B). Thus, the cytoplasmic translocation of Cdc6-wt seems to be mainly regulated by cyclin A-Cdk2–mediated phosphorylation. When we assessed the location of p49-tCdc6 in mock-transfected cells, we found that it occurred in the nucleus in 89.4% of the cells and in the cytoplasm in 10.6% of the cells, indicating that the loss of the NES from the Cdc6-wt suppresses the normal cytoplasmic translocation by 18.9% (Table 1). When cyclin A was coexpressed with p49-tCdc6, there was again a shift to cytoplasmic localization but it was less dramatic than that was observed with Cdc6-wt: 61.5% of the cells showed a nuclear localization (versus 5.7% for Cdc6-wt), whereas 38.5% showed a cytoplasmic localization (versus 94.3% for Cdc6-wt). However, like Cdc6-wt, the cytoplasmic translocation of p49-tCdc6 was almost completely prevented by expressing Cdk2-dn, resulting in a nuclear localization in 99.9% of the transfected cells (Figure 4B and Table 1). Thus, the enhanced cytoplasmic translocation of Cdc6-wt due to up-regulated cyclin A-Cdk2 activity is suppressed by 55.8% when the NES is absent from the protein. Immunoblot analysis showed that Cdc6-wt and p49-tCdc6 exhibit similarly retarded gel mobilities when they are prepared from cells that coexpress cyclin A, whereas these proteins show normal mobility when they are prepared from cells that coexpress Cdk2-dn (Figure 4C). This indicates that Cdc6-wt and p49-tCdc6 are both phosphorylated by cyclin A-Cdk2. Thus, together our data show that although Cdc6-wt and p49-tCdc6 must be phosphorylated before they can be translocated into the cytoplasm, the loss of the NES reduces the efficiency of this phosphorylation-driven translocation.
Figure 4.
The p49-tCdc6 protein is phosphorylated by cyclin A-Cdk2 in HeLa cells, but its cytoplasmic translocation is markedly impaired. Cells were cotransfected with 1.5 μg of pCS2+GFP-mock, 1.5 μg of pCS2+GFP-Cdc6-wt, or 1.5 μg of pCS2+GFP-p49-tCdc6 together with 1.5 μg of pCMV-mock (M), pCMV-cyclin A (CyA), or pCMV-DN-Cdk2 (DN), as indicated. (A) After 24 h, cells were fixed and stained with DAPI. (B) The percentages of transfected cells exhibiting nuclear and cytoplasmic locations were determined; data are expressed as means ± standard deviations from three determinations obtained in each of three experiments. (C) After 24 h, cell extracts were prepared, resolved by 10% SDS-PAGE, and subjected to immunoblot analysis with specific antibodies against Cdc6, cyclin A, Cdk2, and β-actin.
Table 1.
Differential regulation of Cdc6-wt and p49-tCdc6 cytoplasmic translocation by phosphorylation
| Cdc6-wt
|
p49-tCdc6
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Subcellular location(%) | Mock | CyclinA | Cdk-dn | Mock | CyclinA | Cdk-dn | ||||||
| Nucleus | 70.5 | 5.7 | 99.4 | 89.4 | 61.5 | 99.9 | ||||||
| Cytoplasm | 29.5 | 94.3 | 0.6 | 10.6 | 38.5 | 0.1 | ||||||
| Factors contributing to the cytoplasmic translocation of Cdc6 are as follows: | ||||||||||||
| 1) NES: Mock: 29.5% - 10.6% = 18.9%; Cyclin A: 94.3% - 38.5% = 55.8%; Cdk2-dn: No Effect; and | ||||||||||||
| 2) Phosphorylation by cyclin A-Cdk2: Cdc6-wt: 99.4% - 5.7% = 93.7%; p49-tCdc6: 99.9% - 61.5% = 38.4%. | ||||||||||||
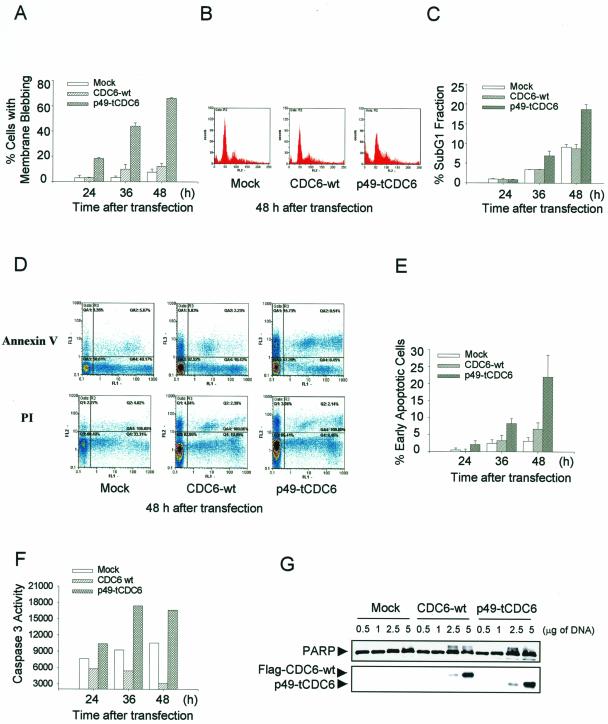
p49-tCdc6 Expression in HeLa Cells Enhances Their Etoposide-induced Apoptosis
To investigate the functional significance of the caspase-3–mediated cleavage of Cdc6 during apoptosis, we asked whether p49-tCdc6 influences the progression of etoposide-induced apoptosis in HeLa cells. Indeed, ectopic expression of the truncated protein but not Cdc6-wt effectively promoted apoptosis in these cells (Figure 5). Expression of p49-tCdc6 caused marked morphological changes, such as membrane blebbing and cell shrinkage, in 80% of the cells, whereas such changes occurred in <20% of cells transfected with either a Cdc6-wt expressing plasmid or vector alone (Figure 5A). Similarly, the rates of DNA condensation and fragmentation, also typical features of apoptotic cells, were markedly increased by the expression of p49-tCdc6 in etoposide-treated cells (Figure 5B). Flow cytometric analysis also showed that the proportion of the subG1 phase in GFP-expressed cells was increased to 16% by the expression of p49-tCdc6 (Figure 5, C and D). To provide further evidence, we also examined the externalization of phosphatidylserine (PS) in p49-tCdc6 expressed cells. Using Alexa 568–labeled annexin-V antibody, we analyzed the percentage of early apoptotic cells in cells expressing the GFP-tagged proteins after treatment with etoposide. The results clearly showed that overexpression of p49-tCdc6 markedly promotes the early apoptotic cell death by three- to fourfold compared with those of the mock and Cdc6-wt transfectants after treatment with etoposide (Figure 5, E and F). Thus, p49-tCdc6, but not Cdc6-wt, promotes apoptosis in etoposide-treated HeLa cells.
Figure 5.
p49-tCdc6 expression in transiently transfected HeLa cells enhances their etoposide-induced apoptosis. HeLa cells were transfected with 3 μg of pCS2+GFP tagged Cdc6-wt and p49-tCdc6 and treated 12 h later with 85 μM etoposide. (A) The percentage of transfected cells showing membrane blebbing was determined 12 h later. Shown are the means ± standard deviations of data from five experiments. (B) Transfected cells were stained with DAPI 12 h after etoposide treatment. The percentage of transfected cells with condensed DNA was quantitated. Shown are the means ± standard deviations of data from five experiments. (C) FACS analysis was carried out with the transfectants 12 h after etoposide treatment. Selective analysis of transfected cells was based on gating a subpopulation of GFP(+) cells as described in MATERIALS AND METHODS. On the graph, X and Y denote green fluorescence intensity (FL1) and propidium iodide intensity (FL2), respectively. The DNA content per cell was determined by staining with propidium iodide. (D) The percentage of cells in the sub-G1 phase was quantitated and represented as a histogram. Apoptotic cells were defined as those with a DNA content lower than that present in the G1 phase. Shown are the means ± standard deviations of data from three experiments. (E) Annexin-V assay was carried out with the transfectants 12 h after etoposide treatment as described in MATERIALS AND METHODS. On the graph, X and Y denote green fluorescence intensity (FL1) and Alexa 568 intensity (FL3) or propidium iodide intensity (FL2), respectively. (F) The percentage of early apoptotic cells was calculated and represented as a histogram. The apoptotic index of the early apoptotic cell populations was calculated by subtracting % PI(+) from % annexin-V(+). Shown are the means ± standard deviations of data from three experiments.
Expression of p49-tCdc6 Induces Apoptosis in HeLa Cells in the Absence of Extracellular Apoptotic Stimuli
We noted that the apoptosis-associated changes in cell morphology, membrane blebbing, and DNA condensation occurred significantly more frequently in untreated control cells expressing p49-tCdc6 than those expressing Cdc6-wt (Figure 5, A and B). We thus examined more closely whether p49-tCdc6 expression can trigger apoptosis in untreated HeLa cells. Surprisingly, cells expressing p49-tCdc6 exhibited all the morphological changes of cells committed to apoptosis, such as cell rounding, shrinkage, and membrane blebbing. The number of cells with membrane blebbing increased by 70% in a time-dependent manner in the 24 to 48 h after transfection. In contrast, cells transfected with the Cdc6-wt expression plasmid or the vector exhibited a minimal increase in the percentage of such cells (Figure 6A). Moreover, FACS analysis showed that the sub-G1 proportion of cells expressing GFP-tagged p49-tCdc6 increased up to 20% in a time-dependent manner over the course of 48 h, whereas the sub-G1 proportion of cells transfected with the Cdc6-wt expression plasmid or the vector was <10% (Figure 6, B and C). In addition, the results from an annexin-V assay also showed that the early apoptotic cell populations increases up to threefold in cells overexpressing p49-tCdc6 compared with the populations in cells overexpressing Cdc6-wt or the mock plasmid (Figure 6, D and E). We further characterized apoptosis-related phenomena in the transfected cells by assessing the degree of PARP cleavage and by measuring caspase-3 activity. The expression of p49-tCdc6 elevated the caspase-3 activation observed 36–48 h after transfection by more than twofold (Figure 6F), whereas these changes were minimal in cells expressing Cdc6-wt and in control cells. Similarly, PARP cleavage was also significantly increased in cells overexpressing p49-tCdc6 (Figure 6G). Thus, the ectopic expression of the p49-tCdc6 protein can induce apoptosis of HeLa cells in the absence of any apoptotic stimuli.
Figure 6.
The Cdc6 cleavage product induces apoptosis in transiently transfected HeLa cells under normal culture conditions. (A) HeLa cells were transfected with 3 μg of pCS2+GFP-Cdc6, and images of live cells were collected by fluorescence microscopy at 24, 36, and 48 h after transfection. The percentage of Cdc6-transfected cells that showed membrane blebbing at these time points was quantitated. (B) HeLa cells were transfected with 3 μg of pCS2+GFP-tagged DNAs. FACS analysis was carried out 48 h after transfection, as described in Figure 5C. (C) The percentage of the cell populations in the sub-G1 phase among GFP(+) cells were calculated and represented as a histogram. (D) Annexin-V assay was carried out with the transfectants expressing GFP in a time-dependent manner, as described in Figure 5E. (E) The percentage of early apoptotic cells was quantitated, as described in Figure 5F. (F) Cell lysates (50 μg of protein) were prepared 24, 36, and 48 h after transfection and incubated with 200 nM Ac-DEVD-AFC in reaction buffer at 37°C for 1 h. The reaction was monitored by fluorescence emission. (G) Nuclear extracts were prepared 36 h after transfection, resolved by 12% SDS-PAGE and subjected to immunoblot analysis by using Cdc6- and PARP-specific antibodies. Shown are the means ± standard deviations of data from three to five experiments.
DISCUSSION
Herein, we show that the Cdc6 protein undergoes a site-specific cleavage mediated by a caspase-3-like protease during the early stages of apoptosis in HeLa cells and SK-HEP-1 cells that have been treated with etoposide, paclitaxel, G-Rh2, or TRAIL. The cleavage of Cdc6 thus seems to be a general phenomenon that occurs in apoptosis. A previous report showed that Cdc6 is proteolytically degraded by a proteasome-dependent pathway during the early stages of programmed cell death induced by the DNA-damaging drug adozelesin or by a distinct caspase-dependent pathway in cells undergoing apoptosis (Blanchard et al., 2002). In this study, we found that Cdc6 is a direct substrate for a caspase-3-like protease, as indicated by the site-specific cleavage of [35S]methionine-labeled Cdc6 by recombinant caspase-3. Furthermore, this cleavage is effectively blocked by a specific caspase-3 inhibitor. Finally, the Cdc6 D442N mutant protein is not cleaved by either recombinant caspase-3 or apoptotic cell extracts (Figure 3), which indicates that the SEVD442/G motif is the caspase-3 cleavage site. This motif loosely conforms to the caspase-3 consensus recognition motif (DXXD↓ followed by a small amino acid residue). Similar caspase-3 cleavage motifs have been identified elsewhere, including SHVD/G, SSTD/S, and SEPD/S in p21-activated kinase 2, prointerleukin-16, and steroid response element-binding proteins, respectively (reviewed in Earnshaw et al., 1999).
Our data are consistent with an earlier report that human Cdc6 is specifically cleaved at SEVD442/G in several human cell lines (HeLa, HL60, and MCF-7) during apoptosis induced by etoposide treatment (Pelizon et al., 2002). This latter report showed that this cleavage generates three products with apparent molecular masses of 52, 45, and 36 kDa and that the cleavage sites for the 52- and 45-kDa products are Asp99 and Asp442, respectively. Herein, we found that cleavage of Cdc6 during apoptosis generated a 49- and a 36-kDa band (Figure 1). The 49-kDa band was uniformly generated in cells undergoing apoptosis induced by a variety of reagents, whose generation kinetics parallels the cleavage of PARP. This band seems to be equivalent to the 45-kDa fragment described previously (Pelizon et al., 2002) because both fragments are generated by cleavage at the Asp442 site. However, we could not detect the 52-kDa fragment observed by Pelizon et al. (2002) under our experimental conditions. This is possibly due to differences in Cdc6 antibodies used. Interestingly, the 36-kDa band was detected only in cells treated with etoposide but not with paclitaxel, G-Rh2, or TRAIL. However, labeled Cdc6 protein is specifically cleaved in vitro into the 49- and 36-kDa fragments by recombinant caspase-3 (Figures 2C and 3B). Therefore, we do not rule out the possibility that the 36-kDa band may not be detectable in some apoptotic cells, possibly because this fragment is highly unstable and is rapidly degraded in these cells. Nevertheless, that Cdc6 is always cleaved into the 49-kDa fragment early in apoptosis induced in different human cell lines by a variety of apoptotic stimuli suggests that this is a crucial event in apoptosis. Supporting this is that the amino acid sequences of the SEVD442/G cleavage site and nearby residues in the human Cdc6 protein are well conserved in the murine Cdc6 protein (Figure 3D), which indicates that the caspase-mediated truncation of Cdc6 and its functional consequences may be conserved among different species.
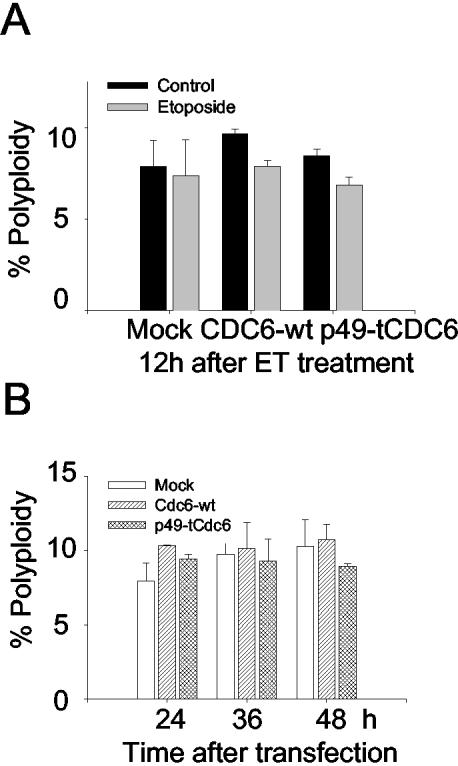
Notably, the N-terminal p49-tCdc6 protein contains almost all the known Cdc6 functional motifs apart from the NES motif. Thus, the p49-tCdc6 protein fragment may be able to translocate into the nucleus and form the prereplication complex but is unable to subsequently translocate to the cytoplasm due to the loss of the NES. Supporting this notion are our experiments examining the cytoplasmic translocation of the Cdc6 protein when the NES is present or absent. We performed these experiments with HeLa cells that have not been treated with apoptotic inducers because the nuclear and cytoplasmic compartments are not well differentiated in shrunken apoptotic cells. We found that the cytoplasmic translocation of Cdc6 is significantly impaired in the absence of the NES even when cyclin A-Cdk2 activity is markedly up-regulated, which normally greatly enhances the cytoplasmic translocation of Cdc6-wt (Table 1 and Figure 4, A and B). In addition, the cytoplasmic translocations of Cdc6-wt and p49-tCdc6 are differentially regulated under the up-regulated conditions of cyclin A-Cdk activity. When cyclin A is expressed and thereby cyclin A-Cdk2 kinase activity is up-regulated in the cells, the cytoplasmic translocation of p49-tCdc6 relative to Cdc6-wt is reduced by 55.8%, compared with 18.9% in the absence of cyclin A expression, consequently resulting in higher rate of the nuclear retention of p49-tCdc6 than that of Cdc6-wt (Table 1). However, when Cdc6 phosphorylation is blocked by the coexpression of the dominant-negative version of Cdk2, the cytoplasmic translocation is almost completely prevented, regardless of whether the NES is present or absent (Table 1 and Figure 4, A and B). These observations reveal several important points. First, the cytoplasmic translocation of Cdc6-wt is mainly regulated by cyclin A-Cdk2–mediated phosphorylation. Second, the loss of the NES suppresses the efficiency of the translocation driven by this phosphorylation. Third, when cyclin A-Cdk2 kinase activity is up-regulated as is during apoptosis, the loss of the NES causes a particularly profound accumulation of the truncated product, even though Cdc6-wt and p49-tCdc6 are equally efficiently phosphorylated (Figure 4C). These results support an earlier report that shows that Cdc6 is released from the prereplicative complex upon phosphorylation by cyclin A-Cdk2 and is then exported to the cytoplasm via its C-terminal NES (Delmolino et al., 2001). Notably, that p49-tCdc6 was still translocated to the cytoplasm, albeit to a lesser extent than the intact protein, strongly suggests that p49-tCdc6 also contains a NES-like sequence. That p49-tCdc6 is generated during early apoptosis and that its cytoplasmic translocation is impaired led us to hypothesize that this phenomenon is functionally linked to the induction of apoptosis. This notion is supported by several observations. First, the caspase-3-mediated cleavage of Cdc6 coincides very well with the onset and continuation of caspase-3 activation and PARP cleavage during apoptosis induced by treatment with different agents (Figure 1). Second, the truncated Cdc6 protein, which contains functional motifs all known to be required for prereplicative complex formation, accumulates in the nucleus and thus could normally form prereplicative complex. The truncated Cdc6 protein in the prereplicative complex could either act as a dominant negative inhibitor of replication, or initiate unscheduled DNA replication. We speculated that any one of these two possibilities could trigger apoptotic cell death. Unscheduled DNA replication is a typical feature of apoptosis induced by DNA-damaging agents (Blanchard et al., 2002). In addition, unscheduled DNA replication can result in polypoidy. For example, when DNA is rereplicated in the absence of an intervening mitotic event or in the absence of a regulating mechanism that prevents reinitiation of replication during the S and G2 phases, polyploidy occurs (reviewed in Kirsch-Volders et al., 1998). Such polyploidy in lymphocytes induced by nocodazole treatment correlates positively with the apoptosis of these cells (Verdoodt et al., 1999). Thus, the caspase-3-mediated truncation of Cdc6 might induce polyploidy and therefore promote apoptosis. Thus, we examined whether overexpression of p49-tCdc6 protein might induce polyploidy in cells treated with or without etoposide. The results from FACS analysis suggest that overexpression of the truncated Cdc6 protein or Cdc6-wt protein does not induce polyploidy and hence not induce reinitiation of unscheduled DNA replication (Figure 7). These results are consistent with earlier studies that showed that misregulation of Cdc6 function is not sufficient to induce overreplication in S. cerevisiae and higher eukaryotes, because overreplication is not observed even when an undegradable mutant Cdc6 protein is overexpressed (reviewed in Pelizon, 2003). These results together with our observations led us to hypothesize that the truncated Cdc6 protein may act as a dominant negative inhibitor of replication, thus promoting apoptotic cell death.
Figure 7.
Overexpression of p49-tCdc6 or Cdc6-wt does not induce polyploidy in HeLa cells treated with or without etoposide. (A) HeLa cells were transfected with 3 μg of pCS2+GFP tagged Cdc6-wt and p49-tCdc6 and then treated for 12 h with 85 μM etoposide. FACS analysis was carried out as described in Figure 5C. The percentages of transfected cells with DNA content >4N were calculated and depicted as a histogram. (B) HeLa cells were transfected with 3 μg of pCS2+GFP-tagged DNAs and trypsinzed at 24, 36, and 48 h after transfection. FACS analysis was carried out as described in Figure 6B. The percentages of transfected cells with DNA content >4N were calculated and represented as a histogram. Shown are the means ± standard deviations of data from three to four experiments.
Our observations strongly support the notion that the nuclear accumulation of p49-tCdc6 is functionally linked to the induction of apoptosis. We found that etoposide-induced apoptosis of cells is markedly increased when p49-tCdc6 is expressed, because there was a fourfold increase in the percentage of cells with membrane blebbing, a twofold increase in the percentage of sub-G1 cells (Figure 5, A, C, and D), and a three- to fourfold increase in annexin-V(+) cell populations compared with those of the mock and Cdc6-wt transfectants after treatment with etoposide (Figure 5, E and F). The results clearly suggest that overexpression of p49-tCdc6, but not of Cdc6-wt, markedly promotes the early apoptotic cell death by three- to fourfold compared with those of the mock and Cdc6-wt transfectants after treatment with etoposide (Figure 5, E and F).
Importantly, the apoptotic progression, as indicated by membrane blebbing (Figure 6A) and the increases in the annexin-V(+) populations and the sub-G1 fraction, is also significantly enhanced in p49-tCdc6–expressing cells that are not treated with etoposide (Figure 6, B–E). Caspase-3 activity and therefore PARP cleavage are also increased in untreated p49-tCdc6-expressing cells (Figure 6, F and G). In contrast, expression of Cdc6-wt or the vector does not significantly induce apoptosis. Thus, p49-tCdc6 indeed can induce apoptosis in the absence of any apoptotic stimuli, indicating that the nuclear accumulation of p49-tCdc6 may play an important role in the induction of apoptosis.
In summary, we propose that the caspase-3–mediated cleavage of Cdc6 regulates the subcellular localization of the protein and that the consequent nuclear accumulation of the p49-tCdc6 protein is functionally linked to apoptosis in cells treated with known inducers of apoptosis.
Acknowledgments
We thank Dr. K.W. Lee (Chungbuk National University, Cheongju, Korea) for comments. This work was supported by grant R01-2000-000-00113-0(2001) from the Korea Research Foundation and by a grant from the National Research Laboratory Fund (M10104000129-02J0000-05910) from the Ministry of Science and Technology, Korea, to S.K.L.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E03–01–0029. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E03-01-0029.
References
- Bermejo, R., Vilaboa, N., and Cales, C. (2002). Regulation of Cdc6, geminin, and Cdt1 in human cells that undergo polyploidization. Mol. Biol. Cell 13, 3989–4000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchard, F., Rusiniak, M.E., Sharma, K., Sun, X., Todorov, I., Castellano, M.M., Gutierrez, C., Baumann, H., and Burhans, W.C. (2002). Targeted destruction of DNA replication protein Cdc6 by cell death pathways in mammals and yeast. Mol. Biol. Cell 13, 1536–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckley, C.D., Pilling, D., Henriquez, N.V., Parsonage, G., Threlfall, K., Scheel-Toellner, D., Simmons, D.L., Akbar, A.N., Lord, J.M., and Salmon, M. (1999). RGD peptides induce apoptosis by direct caspase-3 activation. Nature 397, 534–539. [DOI] [PubMed] [Google Scholar]
- Casenghi, M., Mangiacasale, R., Tuynder, M., Caillet-Fauquet, P., Elhajouji, A., Lavia, P., Mousset, S., Kirsch-Volders, M., and Cundari, E. (1999). p53-independent apoptosis and p53-dependent block of DNA re-replication following mitotic spindle inhibition in human cells. Exp. Cell. Res. 250, 339–350. [DOI] [PubMed] [Google Scholar]
- Castellano, M.M., del-Pozo, J.C., Ramirez-Para, E., Brown, S., and Gutierrez, C. (2001). Expression and stability of Arabidopsis Cdc6 are associated with endoreplication. Plant Cell 13, 2671–2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalton, S., and Whitbread, L. (1995). Cell cycle-regulated nuclear import and export of Cdc47, a protein essential for initiation of DNA replication in budding yeast. Proc. Natl. Acad. Sci. USA 92, 2514–2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delmolino, L.M., Saha, P., and Dutta, A. (2001). Multiple mechanisms regulate subcellular localization of human Cdc6. J. Biol. Chem. 276, 26947–26954. [DOI] [PubMed] [Google Scholar]
- Earnshaw, W.C., Martins, L.M., and Kaufmann, S.H. (1999). Mammalian caspases: structure, activation, substrates, and functions during apoptosis. Annu. Rev. Biochem. 68, 383–424. [DOI] [PubMed] [Google Scholar]
- Guo, M., and Hay, B.A. (1999). Cell proliferation and apoptosis. Curr. Opin. Cell. Biol. 11, 745–752. [DOI] [PubMed] [Google Scholar]
- Hakem, A., Sasaki, T., Kozieradzki, I., and Penninger, J.M. (1999). The cyclin-dependent kinase 2 regulates thymocyte apoptosis. J. Exp. Med. 189, 957–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbig, U., Marlar, C.A., and Fanning, E. (1999). The Cdc6 nucleotide-binding site regulates its activity in DNA replication in human cells. Mol. Biol. Cell 10, 2631–2645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong, F.D., Chen, J., Dinican, S., Schneider, N., and Nisen, P.D. (1999). Taxol, vincristine or nocodazole induces lethality in G1-checkpoint-defective human astrocytoma U353MG cells by triggering hyperploid progression. Carcinogeneis 20, 1161–1168. [DOI] [PubMed] [Google Scholar]
- Jiang, W., Wells, N.J., and Hunter, T. (1999). Multistep regulation of DNA replication by cdk phosphorylation of Hscdc6. Proc. Natl. Acad. Sci. USA 96, 6193–6198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, Y.H., Yoo, K.J., Lee, Y.H., and Lee, S.K. (2000). Caspase 3-mediated cleavage of p21WAF1/CIP1 associated with the cyclin A-cyclin-dependent kinase 2 complex is a prerequisite for apoptosis in SK-HEP-1 cells. J. Biol. Chem. 275, 30256–30263. [DOI] [PubMed] [Google Scholar]
- King, K.L., and Cidlowski, J.A. (1998). Cell cycle regulation and apoptosis. Annu. Rev. Physiol. 60, 601–17. [DOI] [PubMed] [Google Scholar]
- Kirsch-Volders, M., Cundari, E., and Verdoodt, B. (1998). Towards a unifying model for the metaphase/anaphase transition. Mutagenesis 13, 321–335. [DOI] [PubMed] [Google Scholar]
- Lei, M., Kawasaki, Y., and Tye, B.K. (1996). Physical interactions among Mcm proteins and effects of Mcm dosage on DNA replication in Saccharomyces cerevisiae. Mol. Cell. Biol. 16, 5081–5090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levkau, B., Koyama, H., Raines, E.W., Clurman, B.E., Herren, B., Orth, K., Roberts, J.M., and Ross, R. (1998). Cleavage of p21Cip1/Waf1 and p27Kip1 mediates apoptosis in endothelial cells through activation of Cdk 2, role of a caspase cascade. Mol. Cell 1, 553–563. [DOI] [PubMed] [Google Scholar]
- Liang, C., and Stillman, B. (1997). Persistent initiation of DNA replication and chromatin-bound Mcm proteins during the cell cycle in Cdc6 mutants. Genes Dev. 11, 3375–3386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishitani, H., and Nurse, P. (1995). p65cdc18 plays a major role controlling the initiation of DNA replication in fission yeast. Cell 83, 397–405. [DOI] [PubMed] [Google Scholar]
- Park, J.A., Kim, K.W., Kim, S.I., and Lee, S.K. (1998). Caspase 3 specifically cleaves p21WAF1/CIP1 in the earlier stage of apoptosis in SK-HEP-1 human hepatoma cells. Eur. J. Biochem. 257, 242–249. [DOI] [PubMed] [Google Scholar]
- Pelizon, C. (2003). Down to the origin: Cdc6 protien and the competence to replication. Trends Cell Biol. 13, 110–113. [DOI] [PubMed] [Google Scholar]
- Pelizon, C., Fagagna, F.A., Farrace, L., and Laskey, R.A. (2002). Human replication protein Cdc6 is selectively cleaved by caspase-3 during apoptosis. EMBO Rep. 3, 780–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen, B.O., Lukas, J., Sorensen, C.S., Bartek, J., and Helin, K. (1999). Phosphorylation of mammalian Cdc6 by cyclin A/Cdk2 regulates its subcellular localization. EMBO J. 18, 396–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piatti, S., Lengauer, C., and Nasmyth, K. (1995). Cdc6 is an unstable protein whose de novo synthesis in G1 is important for the onset of S phase and for preventing a `reductional' anaphase in the budding yeast Saccharomyces cerevisiae. EMBO J. 14, 3788–3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J., and Russell, D.W. (2001). Molecular Cloning, 3rd ed., Cold Spring Harbor, NY: Cold Spring Harbor Laboratory, 13.31–13.35
- Shi, L., Nishioka, W.K., Th'ng, J., Bradbury, E.M., Litchifield, D.W., and Greenberg, A.H. (1994). Premature p35cdc2 activation is required for apoptosis. Science 263, 1143–1145. [DOI] [PubMed] [Google Scholar]
- Takayama, M., Taira, T., Iguchi-Ariga, S.M.M., and Ariga, H. (2000). Cdc6 interacts with c-Myc to inhibit E-box-dependent transcription by abrogating c-Myc/Max complex. FEBS Lett. 477, 43–48. [DOI] [PubMed] [Google Scholar]
- Takei, Y., Yamamoto, K., and Tsujimoto, G. (1999). Identification of the sequence responsible for the nuclear localization of human cdc6. FEBS Lett. 447, 292–296. [DOI] [PubMed] [Google Scholar]
- Verdoodt, B., Decordier, I., Geleyns, K., Cunha, M., Cundari, E., and Kirsch-Volders, M. (1999). Induction of polyploidy and apoptosis after exposure to high concentrations of the spindle poison nocodazole. Mutagenesis 14, 513–520. [DOI] [PubMed] [Google Scholar]
- Williams, R.S., Shohet, R.V., and Stillman, B. (1997). A human protein related to yeast Cdc6p. Proc. Natl. Acad. Sci. USA 94, 142–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, Z., DeGregori, J., Shohet, R., Leone, G., Stillman, B., Nevins, J.R., and Williams, R.S. (1998). Cdc6 is regulated by E2F and is essential for DNA replication in mammalian cells. Proc. Natl. Acad. Sci. USA 95, 3603–3608. [DOI] [PMC free article] [PubMed] [Google Scholar]