Abstract
In baker's yeast Saccharomyces cerevisiae, cell-cell and cell-surface adhesion are required for haploid invasive growth and diploid pseudohyphal development. These morphogenetic events are induced by starvation for glucose or nitrogen and require the cell surface protein Flo11p. We show that amino acid starvation is a nutritional signal that activates adhesive growth and expression of FLO11 in both haploid and diploid strains in the presence of glucose and ammonium, known suppressors of adhesion. Starvation-induced adhesive growth requires Flo11p and is under control of Gcn2p and Gcn4p, elements of the general amino acid control system. Tpk2p and Flo8p, elements of the cAMP pathway, are also required for activation but not Ste12p and Tec1p, known targets of the mitogen-activated protein kinase cascade. Promoter analysis of FLO11 identifies one upstream activation sequence (UASR) and one repression site (URS) that confer regulation by amino acid starvation. Gcn4p is not required for regulation of the UASR by amino acid starvation, but seems to be indirectly required to overcome the negative effects of the URS on FLO11 transcription. In addition, Gcn4p controls expression of FLO11 by affecting two basal upstream activation sequences (UASB). In summary, our study suggests that amino acid starvation is a nutritional signal that triggers a Gcn4p-controlled signaling pathway, which relieves repression of FLO11 gene expression and induces adhesive growth.
INTRODUCTION
Adherence of cells to one another and to surfaces is a prerequisite for the formation of multicellular structures. In the yeast Saccharomyces cerevisiae, cell-cell and cell-surface adhesion are required for many developmental processes that include mating (Roy et al., 1991; Cappellaro et al., 1994), haploid invasive growth (Roberts and Fink, 1994; Guo et al., 2000), biofilm formation (Reynolds and Fink, 2001), and diploid pseudohyphal development (Gimeno et al., 1992; Mösch and Fink, 1997). Each of these events is initiated by distinct signals that are coupled to the expression of specific cell surface proteins by the corresponding signaling pathways (for recent reviews, see Banuett, 1998; Lengeler et al., 2000). For instance, starvation for glucose causes invasive growth in haploid strains of S. cerevisiae, a developmental event that depends on expression of the cell surface protein Flo11p and that is under control of the mitogen-activated protein kinase (MAPK) and the cAMP pathways (Roberts and Fink, 1994; Mösch et al., 1999; Rupp et al., 1999; Cullen and Sprague, 2000). In diploid strains, starvation for nitrogen induces pseudohyphal growth, a morphogenetic development that also requires Flo11p and functional MAPK and cAMP signaling pathways (Liu et al., 1993; Lo and Dranginis, 1998; Robertson and Fink, 1998; Pan and Heitman, 1999).
FLO11 belongs to a gene family that encodes glycosyl-phosphatidylinositol (GPI)-linked glycoproteins of domain structure similar to the adhesins of pathogenic fungi (Lo and Dranginis, 1996). Flo11p is localized to the cell surface and is required for nutritionally induced cell-cell and cell-surface adhesion during invasive growth, biofilm formation, and pseudohyphal development (Lo and Dranginis, 1998; Guo et al., 2000; Reynolds and Fink, 2001). The unusually large FLO11 promoter is complex and integrates multiple inputs from the cAMP pathway, the MAPK cascade, the mating type, and nutritional signals (Rupp et al., 1999). The transcription factor Flo8p is required for activation of FLO11 by Tpk2p, the catalytic subunit of the cAMP-dependent protein kinase specifically involved in activation of invasive growth and pseudohyphal development (Liu et al., 1996; Pan and Heitman, 2002). Ste12p and Tec1p are transcription factors that are important for FLO11 regulation and transmit signals from the MAPK to sites within the FLO11 promoter that are distinct from the Flo8p target sites (Lo and Dranginis, 1998; Rupp et al., 1999; Köhler et al., 2002). The glucose-responsive protein kinase Snf1p and the transcriptional repressors Nrg1p and Nrg2p also regulate expression of FLO11 (Kuchin et al., 2002).
In S. cerevisiae, starvation for a single amino acid induces a regulatory system known as the general amino acid control (Schürch et al., 1974; Hinnebusch, 1986), which activates transcription of numerous genes encoding enzymes involved in several amino acid biosynthetic pathways (Hinnebusch, 1992), amino acid tRNA synthetases (Meussdoerffer and Fink, 1983; Mirande and Waller, 1988), and enzymes of purine biosynthesis (Mösch et al., 1991). In the general amino acid control system, the sensor kinase Gcn2p phosphorylates the translation initiation factor eIF2 in response to amino acid starvation, an event that results in efficient translation of GCN4 that encodes the transcription factor Gcn4p (Hinnebusch, 1997; Hinnebusch and Natarajan, 2002). Gcn4p activates transcription of target genes by direct promoter binding at sequence-specific Gcn4p-responsive elements (Hope and Struhl, 1985; Oliphant et al., 1989). Two recent studies using genome-wide transcriptional profiling showed that Gcn4p controls expression of >1000 target genes of diverse pathways and functional categories (Jia et al., 2000; Natarajan et al., 2001). These studies demonstrate that Gcn4p has much broader function as a master regulator of gene expression in yeast as previously anticipated. In the human pathogen Candida albicans, Gcn4p has recently been found to coordinate metabolic and morphogenetic reponses to amino acid starvation (Tripathi et al., 2002). However, whether amino acid starvation and Gcn4p in S. cerevisiae or C. albicans control cell-cell and/or cell-substrate adhesion by regulating expression of specific cell-surface proteins, e.g., Flo11p, has not been reported so far.
In this study, we show that amino acid starvation efficiently activates adhesive growth and expression of FLO11 in both haploid and diploid strains in the presence of glucose and ammonium, known suppressors of adhesion. Starvation-induced adhesive growth requires Flo11p and depends on Gcn2p, Gcn4p, Tpk2p, and Flo8p, but not on Ste12p and Tec1p. We find that the FLO11 promoter contains one upstream activation sequence (UASR) and one repression site (URS) that confer regulation by amino acid starvation. Gcn4p seems to be indirectly required to overcome the negative effects of this URS on FLO11 transcription but is not required for regulation of the UASR by amino acid starvation. Gcn4p controls expression of FLO11 by affecting two basal upstream activation sequences (UASB). We suggest that amino acid starvation is a nutritional signal that triggers a Gcn4p-controlled signaling pathway, which relieves repression of FLO11 gene expression and induces adhesive growth.
MATERIALS AND METHODS
Yeast Strains and Growth Conditions
All strains used in this study are derivatives of the S. cerevisiae Σ1278b strain background (Table 1). Deletion mutants for GCN2 (gcn2Δ) were obtained by using the gcn2Δ deletion plasmids pME1658 and pME1659 (Table 2). Plasmids pME1105 and pME1645 were used for constructing gcn4Δ and tpk2Δ mutant strains. Flo8Δ and flo11Δ mutants were obtained by using plasmids pME2155 and pME2156. Transformations were carried out using the lithium-acetate yeast transformation method (Ito et al., 1983). All gene deletions, integrations, or replacements were confirmed by Southern blot analysis (Ausubel et al., 1993). Standard methods for crosses were used and standard yeast culture medium was prepared essentially as described previously (Guthrie and Fink, 1991). For measurements of FLO11 expression, strains were cultivated at 30°C in liquid synthetic minimal medium (YNB) supplemented with 10 mg/l uracile (Ura) and/or 40 mg/l arginine (Arg) where indicated, diluted into fresh medium, and cultivated for 6 h before assaying enzymatic activities or isolation of total RNA. For amino acid starvation, 3-amino-triazole (3AT) or 5-methyl-tryptophan (5MT) was added to cultures at the concentration indicated, and cells were incubated for 8 h before further assays. For nitrogen starvation, cells grown to logarithmic phase were washed with 2% glucose and incubated for 24 h in liquid YNB medium containing 50 μM ammonium sulfate (instead of 50 mM) as the sole nitrogen source. For adhesive growth tests, strains were grown on solid (2% agar) YNB medium containing the indicated supplements and 3AT or 5MT at the respective concentrations to induce amino acid starvation. Qualitative pseudohyphal growth was assayed on SLAD plates (Gimeno et al., 1992).
Table 1.
Yeast strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| RH2648 | MATaura3-52 | This study |
| RH2649 | MATagcn2 Δ::LEU2 ura3-52 leu2::hisG | This study |
| RH2650 | MATagcn4 Δ::LEU2 ura3-52 leu2::hisG | This study |
| RH2651 | MATagcn4 Δ::LEU2 gcn2Δ::kanR ura3-52 leu2::hisG | This study |
| RH2652 | MATaflo8Δ::kanR ura3-52 | This study |
| RH2653 | MATaflo8Δ::kanR gcn2Δ::LEU2 ura3-52 leu2::hisG | This study |
| RH2654 | MATaflo8 Δ::kanR gcn4 Δ::LEU2 ura3-52 leu2::hisG | This study |
| RH2656 | MATa/α ura3-52/ura3-52 trp1::hisG/TRP1 | This study |
| RH2657 | MATa/α gcn2Δ::LEU2/gcn2Δ::LEU2 ura3-52/ura3-52 leu2::hisG/leu2::hisG trp1::hisG/TRP1 | This study |
| RH2658 | MATa/α gcn4Δ::LEU2/gcn4Δ::LEU2 ura3-52/ura3-52 leu2::hisG/leu2::hisG trp1::hisG/TRP1 | This study |
| RH2659 | MATa/α tpk2Δ::kanR/tpk2Δ::kanR ura3-52/ura3-52 leu2::hisG/LEU2 his3::hisG/HIS3 trp1::hisG/TRP1 | This study |
| L5627 | MATa/α ste12Δ::LEU2 ura3-52/ura3-52 leu2::hisG/leu2::hisG | Liu et al., (1993) |
| HMC267 | MATa/α tec1::LEU2/tec1::LEU2 ura3-52/ura3-52 leu2::hisG/leu2::hisG trp1::hisG/TRP1 | Mösch and Fink (1997) |
| RH2660 | MATa/α flo8Δ::kanR/flo8Δ::kanR ura3-52/ura3-52 trp1::hisG/TRP1 | This study |
| RH2661 | MATa/α flo11Δ::kanR/flo11Δ::kanR ura3-52/ura3-52 trp1::hisG/TRP1 | This study |
| RH2662 | MATaflo11Δ::kanR ura3-52 | This study |
Table 2.
Plasmids used in this study
| Plasmid | Description | Source |
|---|---|---|
| pME1092 | 2.8-kb fragment containing GCN4 in pRS314 | Albrecht et al. (1998) |
| pME1098 | 2.8-kb fragment containing GCN4m in pRS314 | Albrecht et al. (1998) |
| pME1105 | Cassette for full deletion of GCN4-open reading frame (LEU2) | Albrecht et al. (1998) |
| pME1658 | Cassette for full deletion of GCN2-open reading frame (LEU2) | Grundmann et al. (2001) |
| pME1659 | Cassette for full deletion of GCN2-open reading frame (kanr) | Grundmann et al. (2001) |
| pME2155 | Cassette for full deletion of FLO8-open reading frame (kanr) | This study |
| pME2156 | Cassette for full deletion of FLO11-open reading frame (kanr) | This study |
| pME1645 | Cassette for full deletion of TPK2-open reading frame (kanr) | This study |
| pME1765 | pBluescriptKS+ containing kanr-cassette | Grundmann et al. (2001) |
| pME2519 | 7.43-kb fragment containing FLO11 in YCplac33 | This study |
| B3782 | 3-kb FLO11-promoter fragment in YEp355 | Rupp et al. (1999) |
| pflo11-1 to pflo11-15 | 200 bp deletions in B3782 from-1 to -201 bp, -202 to -401 bp until -2802 to -3001 bp | Rupp et al. (1999) |
| pME2212 | pLG669Z-UASΔ | This study |
| pFLO11-2/1 to pFLO11-15/14 | 440 bp FLO11 promoter sequence elements cloned into pLG669Z | Rupp et al. (1999) |
| pFLO11-5 | 239-bp FLO11 promoter element cloned into pLG669Z | This study |
| pFLO11-6 | 239-bp FLO11 promoter element cloned into pLG669Z | This study |
| pFLO11-9 | 247-bp FLO11 promoter element cloned into pLG669Z | This study |
| pFLO11-10 | 239-bp FLO11 promoter element cloned into pLG669Z | This study |
Plasmids
Plasmids used in this study are listed in Table 2. Plasmid pME2155 carrying the flo8Δ:kanR deletion cassette was constructed by amplifying the plasmid backbone and sequences flanking the FLO8 open reading frame from plasmid pHL129 (Liu et al., 1996) by using the two primers OG33 (5′-GAAGATCTTCTACCACGGAATGCGTTTCC-3′) and OG34 (5′-GAAGATCTCTGACATTTCGCTAAATTTGGG-3′) to create a BglII restriction site, which was used to insert the kanR kanamycin resistance cassette of pME1765 (Grundmann et al., 2001). Similarly, deletion cassettes for FLO11 (pME2156) and TPK2 (pME1645) were created by replacement of the FLO11 or TPK2 open reading frames by kanR as selectable marker. Plasmid pME2519 carrying a functional FLO11 gene was constructed by amplification of FLO11 as an EcoRI-SalI fragment (sequence number relative to the initiating AUG: –3043 to +4392) by using polymerase chain reaction (PCR) and primers FLO11-102 (5′-CCGGAATTCGTGGCGCGGTGCCAATACTACCGGTACTTG-3′) and FLO11-103 (5′-ACGCGTCGACCCCCAATTCAAGAATACAATTACTTAGCGTGG-3′) and insertion of the fragment into the EcoRI and SalI sites of plasmid YC-plac33 (Gietz and Sugino, 1988). Plasmid pME2212 was obtained by deletion of the 434-base pair XhoI fragment containing the endogenous UAS element of the CYC1-promotor region in plasmid pLG669Z (Guarente and Ptashne, 1981). To obtain plasmids pFLO11-5, pFLO11-6, pFLO11-9, and pFLO11-10, individual FLO11 promoter fragments were amplified by PCR and cloned into pME2212 by using a restriction site (XhoI) introduced at the 5′ end of the PCR primers. The primers used have been described previously (Rupp et al., 1999) and were #5F and #5R to obtain pFLO11-5, #6F and #6R for pFLO11-6, #9F and #9R for pFLO11-9, and #10F and #10R for pFLO11-10.
Northern Hybridization Analysis
Total RNAs from yeast were isolated essentially as described previously from cultures grown in liquid media (Cross and Tinkelenberg, 1991). RNAs were separated on 1.4% agarose gel containing 3% formaldehyde and transferred onto nylon membranes by electroblotting. Gene specific probes were 32P-radiolabeled with the HexaLable DNA labeling kit (MBI Fermentas, St. Leon-Rot, Germany). Hybridizing signals were quantified using a BAS-1500 phosphorimaging scanner (Fuji, Tokyo, Japan).
β-Galactosidase Assay
Assays were performed with extracts of cultures grown in liquid media. Specific β-galactosidase activity was normalized to the total protein (Bradford, 1976) in each extract and equalized (OD415 × 1.7)/(0.0045 × protein concentration × extract volume × time) (Rose and Botstein, 1983). Assays were performed for at least three independent transformants, and the mean value is presented. The SEs of the means were <15%.
Growth Tests and Photomicroscopy
Adhesive growth tests with haploid and diploid strains were performed essentially as described previously (Roberts and Fink, 1994). Strains were pregrown on solid YNB medium containing the indicated supplements for 20 h. Cells were then patched on fresh YNB containing supplements and 3AT or 5MT at the indicated concentrations and incubated at 30°C for 1 to 5 days. Plates were photographed and then carefully washed under a stream of water. The plates were photographed once again to document the remaining adhesive cells. Pseudohyphal growth was viewed with an Axiovert microscope (Carl Zeiss, Jena, Germany) and photographed using a Xillix microimager digital camera with the Improvision Openlab software (Improvision, Coventry, United Kingdom).
RESULTS
Amino Acid Starvation Activates Adhesive Growth and FLO11 Gene Expression in the Presence of Glucose and Ammonium in S. cerevisiae
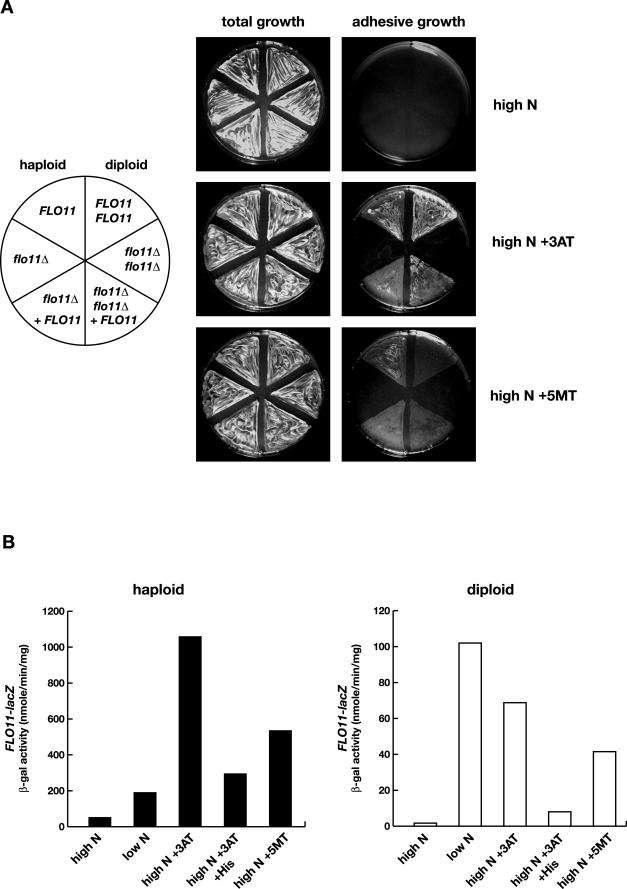
We tested, whether starvation for amino acids activates adhesive growth and expression of FLO11 in yeast. Haploid and diploid wild-type and flo11Δ mutant strains were tested for adhesive growth on solid high ammonium medium with or without the addition of 3AT, a histidine-analog that induces histidine starvation (Hilton et al., 1965; Schürch et al., 1974). Haploid strains are known to exhibit adhesive growth on high ammonium medium, but only after prolonged incubation of 4–5 d (Roberts and Fink, 1994). Therefore, we assayed invasive growth after 24 h, a time period that is not sufficient to induce substrate adhesion in haploids (Lo and Dranginis, 1996; Rupp et al., 1999). As expected, haploid cells did not adhere significantly to the agar substrate after 24 h of growth on nonstarvation medium (Figure 1A) but became adhesive after prolonged incubation of 5 d (Figure 3A). In contrast, haploid cells became highly adhesive already after 24 h of growth on amino acid starvation medium (Figure 1A). Adhesive growth was dependent on FLO11, because a flo11Δ mutant strain was nonadhesive under all conditions tested, and adhesive growth on starvation medium could be restored by complementation of the flo11Δ mutant with a functional FLO11 gene (Figure 1A). Similar results were obtained for diploid strains. Diploid strains were nonadhesive under nonstarvation conditions (even after 5 d), but they became highly adhesive when starved for amino acids even in the presence of high amounts of ammonium (Figures 1A and 4A). Deletion of FLO11 blocked diploid adhesive growth under amino acid starvation conditions, and adhesion of a diploid flo11Δ/flo11Δ mutant was restored by complementation with FLO11 on a plasmid (Figure 1A).
Figure 1.
Amino acid starvation induces adhesive growth and FLO11 expression in haploid and diploid S. cereivisiae strains. (A) Haploid strains RH2648 (FLO11) and RH2662 (flo11Δ), and diploid strains RH2656 (FLO11/FLO11) and RH2661 (flo11Δ/flo11Δ) carrying plasmid B3782 or pME2519 (+FLO11) were patched on YNB (high N), YNB + 5 mM 3AT (high N + 3AT) or YNB + 1 mM 5MT (high N + 5MT). Plates were incubated at 30°C for 24 h and photographed before (total growth) and after (adhesive growth) nonadhesive yeast cells were washed off the agar surface. (B) Expression of the FLO11-lacZ reporter gene was determined in yeast strains RH2648 (haploid) and RH2656 (haploid) carrying plasmid B3782 under different nutritional conditions. Cultures grown in YNB were used for assaying high ammonium conditions (high N, 50 mM ammonium sulfate). Nitrogen starvation (low N) was induced by growth in YNB with limited amounts of ammonium sulfate (50 μM) as sole nitrogen source. Amino acid starvation was induced by addition of 10 mM 3AT (high N + 3AT) or 1 mM 5MT (high N + 5MT). To revert histidine starvation, 1 mM histidine was added to YNB medium containing 10 mM 3AT (high N + 3AT +His). Units of specific β-galactosidase activities are shown in nanomoles per minute per milligram. Bars depict means of at least three independent measurements with a SD not exceeding 15%.
Figure 3.
Requirement of GCN2, GCN4, and FLO8 for amino acid starvation-induced adhesive growth and FLO11 expression in haploid S. cerevisiae strains. (A) Haploid yeast strains RH2648 (control), RH2649 (gcn2Δ), RH2650 (gcn4Δ), RH2651 (gcn2Δ gcn4Δ), RH2662 (flo11Δ), RH2652 (flo8Δ), RH2653 (flo8Δ gcn2Δ) and RH2654 (flo8Δ gcn4Δ) carrying plasmid B3782 were patched on YNB +Arg medium (high N) and on YNB + Arg containing either 1 mM 3AT (high N + 1 mM 3AT) or 10 mM 3AT (high N + 10 mM 3AT). Plates were incubated at 30°C for 1 d (3AT-containing media) or 5 d (nonstarvation medium) and photographed before (total growth) and after (adhesive growth) nonadhesive cells were washed off the agar surface. (B) Expression of FLO11-lacZ. Strains used in A were grown to logarithmic phase in YNB +Arg medium (high N) or YNB +Arg medium containing 10 mM 3AT (high N + 10 mM 3AT) before specific β-galactosidase activities were measured. Bars depict means of at least three independent measurements of β-galactosidase activities with a SD not exceeding 15%. Units are nanomoles per minute per milligram.
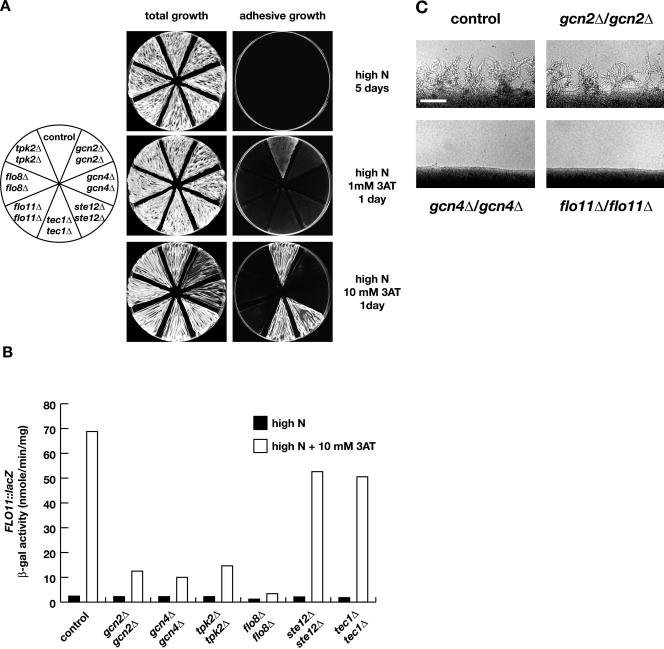
Figure 4.
Requirement of GCN2, GCN4, FLO8, and TPK2 for amino acid starvation-induced adhesive growth and FLO11 expression in diploid S. cerevisiae strains. (A) Diploid yeast strains RH2656 (control), RH2657 (gcn2Δ/gcn2Δ), RH2658 (gcn4Δ/gcn4Δ), RH2659 (tpk2Δ/tpk2Δ), RH2660 (flo8Δ/flo8Δ), RH2661 (flo11Δ/flo11Δ), L5627 (ste12Δ/ste12Δ), and HMC267 (tec1Δ/tec1Δ) carrying plasmid B3782 were assayed for adhesive growth as described for haploid strains in Figure 3. (B) Expression of FLO11-lacZ. Strains used in A were grown to logarithmic phase in YNB + Arg medium (high N) or YNB + Arg medium containing 10 mM 3AT (high N + 10 mM 3AT) before specific β-galactosidase activities were measured. Bars depict means of at least three independent measurements of β-galactosidase activities with a SD not exceeding 15%. Units are nanomoles per minute per milligram. (C) Diploid yeast strains RH2656 (control), RH2657 (gcn2Δ/gcn2Δ), RH2658 (gcn4Δ/gcn4Δ), and RH2661 (flo11Δ/flo11Δ) carrying plasmid B3782 were streaked on nitrogen starvation plates (SLAD) for induction of pseudohyphal growth. Pictures weretakenafter 3 d of incubation at 30°C. Bar, 50 μm.
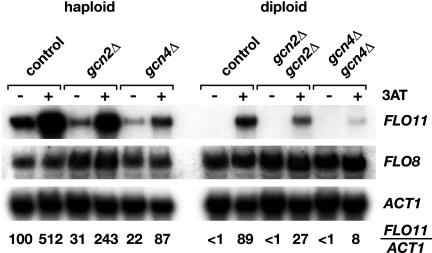
Expression of FLO11 was measured in haploid and diploid yeast strains under different nutritional conditions, to determine the correlation between adhesive growth and expression of FLO11. When using a FLO11-lacZ reporter gene, a 3.6-fold increase in expression was found in nitrogen-starved haploids compared with nonstarved cells, and a 59-fold increase was measured in diploid cells (Figure 1B). An induction of FLO11 expression by nitrogen starvation has been observed previously (Lo and Dranginis, 1998; Rupp et al., 1999). Herein, we found that amino acid starvation led to an increase in the expression of FLO11-lacZ of 20-fold in haploid cells and 41-fold in diploid cells even when high amounts of ammonium are available (Figure 1B). Induction of FLO11-lacZ expression by the histidine-analog 3AT was partially reversible by addition of histidine (Figure 1B), suggesting that histidine starvation is the inducing signal for enhanced expression. The effect of amino acid starvation on transcript levels of FLO11 was determined, to corroborate the data obtained with the FLO11-lacZ translational fusion. In haploid strains, amino acid starvation in the presence of high amounts of ammonium led to a 5.1-fold increase in FLO11 transcript levels (Figure 2), correlating with FLO11-lacZ expression. In nonstarved diploid cells, very low FLO11 transcripts were detectable in Northern hybridization experiments (Figure 2). In 3AT-treated diploid cells, the amount of FLO11 transcripts increased to a level comparable to that found in nonstarved haploids (Figure 2), thus correlating with FLO11-lacZ expression. These results show that amino acid starvation not only enhances adhesive growth of haploid and diploid cells in a FLO11-dependent manner but also causes a strong increase in the expression of FLO11 in both cell types. However, FLO11 expression does not seem to strictly correlate with adhesive growth, because nonstarved haploids are comparable with starved diploids with respect to FLO11 transcript levels and expression of FLO11-lacZ, but not with respect to adhesive growth. This suggests that amino acid starvation-induced adhesive growth not only involves up-regulation of FLO11 expression but also additional factors whose functions depend on Flo11p.
Figure 2.
Transcript levels of FLO11, FLO8, and ACT1. Total RNAs were prepared from haploid yeast strains RH2648 (control), RH2649 (gcn2Δ), RH2650 (gcn4Δ), and the diploid yeast strains RH2656 (control), RH2657 (gcn2Δ/gcn2Δ), RH2658 (gcn4Δ/gcn4Δ) grown in high ammonium medium (YNB + Ura + Arg) to logarithmic phase before (–) and after (+) induction of amino acid starvation by addition of 10 mM 3AT. For measurements of FLO11, FLO8, and ACT1 transcript levels, 10 μg of total RNA from each sample was subjected to a Northern hybridization analysis. Signals were quantified using a phosphorimaging scanner. Numbers given indicate relative expression levels of FLO11 when compared with ACT1 as internal standard with a value for nonstarvation expression of the haploid control strain set to 100. Values represent the average of three independent measurements.
The tryptophan-derivative 5MT is a further amino acid analog known to induce amino acid starvation (Schürch et al., 1974). We also determined the effects of adding 5MT to high ammonium medium on adhesive growth and expression of FLO11. Results obtained by addition of 5MT were similar to that obtained by adding 3AT, although effects were less pronounced (Figure 1).
Haploid Adhesive Growth and Expression of FLO11 Depend on GCN2 and GCN4, Elements of the General Amino Acid Control System
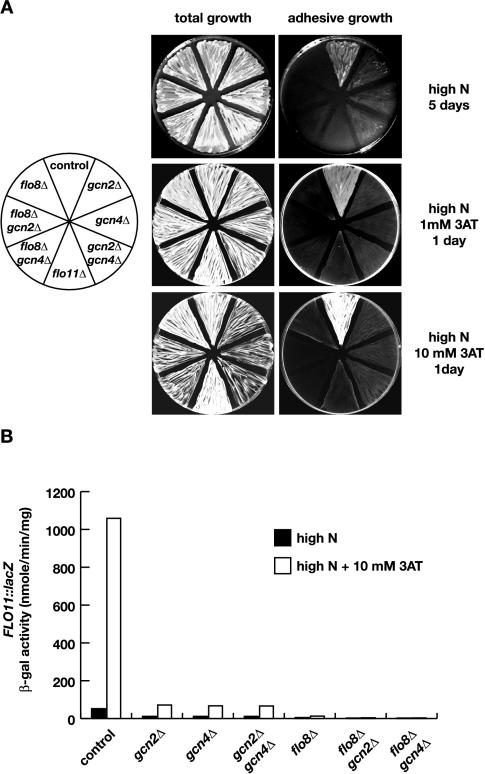
The requirement of the sensor kinase Gcn2p and the transcriptional activator Gcn4p for adhesive growth and expression of FLO11 was analyzed in haploid gcn2Δ, gcn4Δ, and gcn2Δ gcn4Δ mutant strains and compared with a flo11Δ strain. Under nonstarvation conditions, adhesive growth of haploid strains was significantly reduced in the absence of GCN2 or GCN4 (Figure 3A). The gcn2Δ gcn4Δ double mutant was indistinguishable from the single mutants. Amino acid starvation was induced by addition of either 1 mM 3AT or 10 mM 3AT to the growth medium, because addition of 10 mM 3AT inhibits growth of strains lacking GCN2 or GCN4 (Figure 3A). Addition of 1 mM 3AT was sufficient to enhance adhesive growth of a control strain, without signifi-cantly inhibiting growth of the gcn2Δ or gcn4Δ mutant strains (Figure 3A). Adhesive growth of haploid gcn2Δ, gcn4Δ, or gcn2Δ gcn4Δ mutants was reduced under both 1 mM and 10 mM 3AT starvation conditions, compared with the control strain.
Strains measured for adhesive growth were further assayed for expression of the FLO11-lacZ reporter gene and FLO11 transcript levels. Under nonstarvation conditions, expression of FLO11-lacZ dropped from 52.1 U in a control strain to 11.5 U in the gcn2Δ, 11.5 U in the gcn4Δ, and 11.4 U in the gcn2Δ gcn4Δ double mutant strain, respectively (Figure 3B). The decrease in FLO11-lacZ expression in the gcn mutant strains correlated with a decrease in both FLO11 transcript levels (Figure 2) and the resulting adhesive growth behavior (Figure 3A), although deletion of GCN2 led to a less severe reduction of FLO11 mRNA than observed for strains lacking GCN4. To induce amino acid starvation, 3AT was added to strains grown to logarithmic phase at concentrations of both 1 and 10 mM. Addition of 1 mM 3AT was sufficient to induce expression of FLO11-lacZ to levels (683 U) almost matching that obtained by the addition of 10 mM 3AT (1059 U) (Figure 3B). Expression of FLO11-lacZ signifi-cantly decreased by deletion of GCN2 (71.5 U), GCN4 (67.3 U), or both (66.5 U) under 10 mM 3AT conditions, corresponding to a roughly 16-fold drop in comparison to the expression measured in the control strain. These data suggest that Gcn2p and Gcn4p are required for full induction of FLO11 expression by amino acid starvation. However, FLO11-lacZ expression and FLO11 transcript levels measured in the starved gcn mutant strains were similar to levels measured in a control strain under nonstarvation conditions. Yet these strains were less adhesive under starvation conditions (Figure 3). This result indicates that Gcn2p and Gcn4p not only control expression of the FLO11 gene but also might regulate Flo11p at a posttranslational level or expression of additional genes required for adhesive growth.
We further tested, whether high-level expression of Gcn4p is sufficient to induce adhesive growth and enhanced expression of FLO11. For this purpose, a mutant allele of GCN4 (GCN4m) was expressed that carries point mutations inactivating all four μ open reading frames in the GCN4 upstream leader and causes high expression of Gcn4p under nonstarvation conditions (Mueller and Hinnebusch, 1986). However, expression of GCN4m was not sufficient to induce adhesive growth and did not lead to enhanced expression of FLO11-lacZ (our unpublished data), indicating that Gcn4p might control expression of FLO11 in concert with other transcriptional regulators or by an indirect mechanism.
In summary, our results show that Gcn2p and Gcn4p, elements of the general control system of amino acid biosynthesis, are required for adhesive growth and efficient expression of FLO11 in haploid cells.
In Diploid Yeast Cells, GCN4 Is Required for Amino Acid Starvation-induced Adhesive Growth and Nitrogen Starvation-induced Pseudohyphal Development
Amino acid starvation-induced adhesive growth, FLO11-lacZ expression and FLO11 transcript levels were further measured in diploid strains. Under nonstarvation conditions, diploid gcn2Δ/gcn2Δ and gcn4Δ/gcn4Δ mutant strains were indistinguishable from a control strain with respect to their nonadhesive growth behavior and the low expression of FLO11-lacZ or FLO11 transcripts (Figures 2 and 4). When starved for amino acids, deletion of either GCN2 or GCN4 significantly suppressed the adhesive growth, which was induced in the control strain. This finding correlated with a decrease in expression of FLO11-lacZ of 4.6-fold in the gcn2Δ/gcn2Δ strain (15 U) and of 5.5-fold in the gcn4Δ/gcn4Δ mutant (12.5 U) in comparison with the induced levels measured in the control strain (Figure 4, A and B). Concomitantly, FLO11 transcript levels decreased 3.3-fold in the absence of GCN2 in comparison with the control strain and 11-fold, when GCN4 was deleted. Thus, amino acid starvation-induced adhesive growth and expression of FLO11 require GCN2 and GCN4 in diploid strains, corroborating the data obtained in haploids.
Cell-cell and cell-substrate adhesion are processes essential for the development of pseudohyphal filaments of diploid S. cerevisiae strains that have been starved for nitrogen (Lo and Dranginis, 1998). Diploid strains lacking GCN2 or GCN4 were tested for pseudohyphal development on nitrogen starvation medium. Only gcn4Δ/gcn4Δ mutant strains were suppressed for development of pseudohyphae comparable with flo11Δ/flo11Δ mutant strains (Figure 4C).
In summary, Gcn2p and Gcn4p are required for amino acid starvation-induced adhesive growth in diploids, but for pseudohyphal development, induced by nitrogen starvation, only Gcn4p is necessary.
Amino Acid Starvation-induced Expression of FLO11 Requires the Transcription Factors Gcn4p and Flo8p, but Not Ste12p and Tec1p
Adhesive growth and expression of FLO11 are under control of the cAMP pathway and the MAPK pathway (Pan and Heitman, 1999; Rupp et al., 1999). We tested whether amino acid starvation-induced adhesive growth and expression of FLO11 requires Tpk2p or Flo8p, elements of the cAMP-regulated pathway, or the transcription factors Ste12p and Tec1p, elements of the MAPK pathway. Adhesive growth of diploid strains lacking TPK2 or FLO8 was suppressed to a degree comparable with a flo11Δ/flo11Δ control strain under amino acid starvation conditions (Figure 4A). Expression of FLO11-lacZ was reduced 4.3-fold in tpk2Δ/tpk2Δ strains (16 U) and 20-fold in flo8Δ/flo8Δ mutants (3.4 U). In comparison, deletion of STE12 or TEC1 did not suppress adhesive growth in the presence of 10 mM 3AT, and expression of FLO11-lacZ was reduced only 1.4-fold in both the ste12Δ/ste12Δ (52 U) and tec1Δ/tec1Δ (49 U) mutant strains (Figure 4B). Thus, efficient expression of FLO11 under amino acid starvation conditions requires FLO8 and TPK2, but not STE12 and TEC1.
3AT-induced adhesive growth and expression of FLO11-lacZ was measured in haploid flo8Δ gcn2Δ and flo8Δ gcn4Δ double mutant strains and compared with gcn2Δ, gcn4Δ, and flo8Δ single mutants, to distinguish between a parallel and a linear configuration of the general control system and the transcription factor Flo8p. Under amino acid starvation conditions, both adhesive growth and expression of FLO11-lacZ is lower in the flo8Δ gcn2Δ (4 U) and flo8Δ gcn4Δ (2.9 U) double mutants than in the gcn2Δ (71.5 U), gcn4Δ (67.3 U), or flo8Δ (13 U) single mutants (Figure 3). The additive effects of the gcn2Δ and flo8Δ or gcn4Δ and flo8Δ mutations argue for independent functions of the general control system and Flo8p. This conclusion is supported by the fact that transcript levels of FLO8 are not affected by amino acid starvation or by mutations in GCN2 or GCN4 (Figure 2).
In summary, amino acid starvation-induced expression of FLO11 requires the combined action of the transcription factors Gcn4p and Flo8p but does not depend on Ste12p and Tec1p.
Identification of FLO11 Promoter Elements Mediating Regulation by Amino Acid Starvation
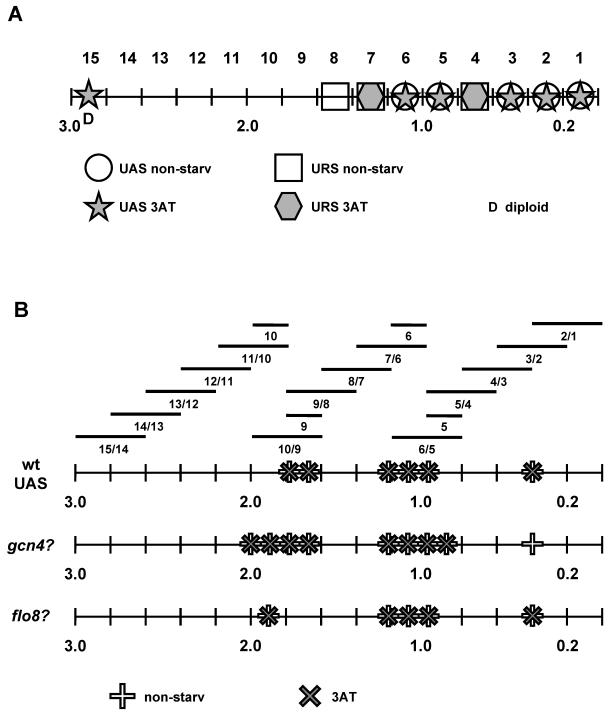
A set of 15 flo11-lacZ promoter deletion constructs spanning the region between the 3000 base pairs upstream of the FLO11 initiation codon was used (Rupp et al., 1999) to identify FLO11 promoter elements that confer regulation of FLO11 expression in response to amino acid starvation. Expression of this set of flo11-lacZ reporter constructs, each containing an individual 200-base pair deletion, was assayed in haploid and diploid strains under both nonstarvation and amino acid starvation conditions and compared with the intact FLO11-lacZ reporter (Table 3). A deletion was assigned to contain a UAS (upstream activation site) when leading to at least 50% reduced expression of FLO11-lacZ, and as a URS (upstream repression site) when causing at least threefold enhanced expression (Figure 5A).
Table 3.
Expression of different FLO11-lacZ reporter constructs under nonstarvation and amino acid starvation conditions in haploid and diploid S. cerevisiae strains
|
MATa
|
MATa/α
|
||||||
|---|---|---|---|---|---|---|---|
| Construct | Boundaries of deletion | YNB | YNB +3AT | 3AT induction | YNB | YNB +3AT | 3AT induction |
| FLO11 | —none | 52.1 (11.5) | 1059 (67.3) | 20.3 (5.9) | 1.7 (1.9) | 68.8 (12.5) | 40.5 (6.9) |
| flo11-Δ1 | -201 to -1 | 2.2 | 39.8 | 17.8 | 0.1 | 1.1 | 11.0 |
| flo11-Δ2 | -401 to -202 | 13.7 | 284 | 20.7 | 0.7 | 7.8 | 11.1 |
| flo11-Δ3 | -600 to -401 | 13.7 | 299 | 21.9 | 0.6 | 6.4 | 10.6 |
| flo11-Δ4 | -800 to -601 | 197 | 1693 | 8.6 | 5.7 | 208 | 36.5 |
| flo11-Δ5 | -1000 to -801 | 0.3 | 0.2 | 0.7 | 0.1 | 0.4 | 4.0 |
| flo11-Δ6 | -1200 to -1001 | 1.9 | 7.0 | 3.6 | 0.6 | 1.6 | 2.7 |
| flo11-Δ7 | -1400 to -1201 | 363 (5.2) | 4597 (62.2) | 12.7 (12.1) | 13.0 (3.8) | 380 (30.5) | 29.2 (8.0) |
| flo11-Δ8 | -1600 to -1401 | 375 (680) | 397 (850) | 1.1 (1.3) | 132 (166) | 98.0 (297) | 0.7 (1.8) |
| flo11-Δ9 | -1800 to -1601 | 30.8 (17.5) | 680 (18.2) | 22.1 (1.0) | 1.0 (1.1) | 28.2 (3.1) | 38.2 (2.8) |
| flo11-Δ10 | -2000 to -1801 | 51.4 | 1035 | 20.1 | 0.8 | 36.1 | 45.1 |
| flo11-Δ11 | -2200 to -2001 | 63.4 | 1328 | 20.9 | 0.8 | 54.2 | 67.8 |
| flo11-Δ12 | -2400 to -2201 | 65.4 | 1215 | 18.6 | 1.1 | 41.2 | 37.5 |
| flo11-Δ13 | -2600 to -2401 | 49.1 | 1003 | 20.4 | 1.2 | 54.0 | 45.0 |
| flo11-Δ14 | -2800 to -2601 | 108 | 1312 | 12.2 | 2.1 | 81.4 | 38.8 |
| flo11-Δ15 | -3000 to -2801 | 38.1 | 910 | 23.9 | 1.1 | 16.1 | 14.6 |
Expression of indicated FLO11-lacZ reporter constructs (Rupp et al., 1999) was assayed in the haploid strain RH2648 (MATa) and the diploid strain RH2656 (MATa/α) under nonstarvation conditions (YNB + Arg) and under amino acid starvation conditions (YNB + Arg +3AT). Values shown in parentheses were obtained by assaying constructs in gcn4Δ strains RH2650 (MATa gcn4Δ) and RH2658 (MATa/α gcn4Δ/gcn4Δ), respectively. Deleted segments in the individual constructs are numbered according to the numbering shown in Figure 5. Boundaries of deletions are indicated with respect to the translational start site of FLO11 at position +1. Given numbers represent β-galactosidase activities in actual units nanomoles per minute per milligram. Values for 3AT induction represent the ratio of activities obtained for a given FLO11-lacZ construct in the presence or absence of 3AT, respectively. All values are means of at least three independent measurements with a standard deviation not exceeding 15%.
Figure 5.
Sequence elements involved in regulation of FLO11. (A) The specific β-galactosidase activities of 15 yeast strains carrying individual 200-base pair deletions of the FLO11 promoter region were compared with a yeast strain carrying the full-length 3-kbp wild-type promoter under nonstarvation (nonstarv) and amino acid starvation conditions (3AT) in haploid as well as in diploid yeasts (Table 3). Individual 200-base pair segments within the FLO11 promoter region are numbered from 1 to15 and their relative position in kilobases is shown with respect to the translational start site. A ratio of specific β-galactosidase activity (deletion/3 kbp promoter) of three or higher was defined as URS. URS elements under nonstarvation conditions are marked by a square (URS nonstarv) and under starvation conditions by a hexagon (URS 3AT). At least 50% lower expression than the wild-type reporter is represented as an UAS. UAS elements under nonstarvation conditions are marked by a circle (UAS nonstarv) and under starvation conditions by a star (UAS 3AT). Elements, which occur only in the diploid strain are marked by a D. (B) Expression of 19 CYC1-lacZ reporter constructs carrying isolated segments of the FLO11 promoter region was determined in haploid wild-type cells or in corresponding cells deleted for the transcription factors GCN4 or FLO8 under nonstarvation or amino acid starvation conditions (Table 4). The symbols (white cross, nonstarvation conditions, gray cross, starvation conditions) are placed on a line in a position that indicates, which of the fragments stimulated β-galactosidase activity. Each line represents the FLO11 promoter in the indicated genetic background. The first row (wt UAS) denotes sequence elements showing a >5-fold elevation of the reporter over a plasmid without an insert. The next two lines (gcn4Δ and flo8Δ) represent sequence elements showing a >3-fold reduction of the β-galactosidase activity in the mutant (gcn4Δ or flo8Δ) compared with the activity of the element in the control strain.
The flo11-1, flo11-2, flo11-3, flo11-5, and flo11-6 deletions identify at least five UAS elements. The UAS sites deleted in flo11-1, flo11-2, and flo11-3 do not seem to confer amino acid starvation signals for the expression of FLO11, because their absence does not suppress induction of FLO11 expression by 3AT in haploids. In contrast, the UAS sites defined by flo11-5 and flo11-6 might be involved in amino acid starvation-induced activation, because their deletion drastically reduced activation by 3AT in haploids and diploids. However, these UAS elements presumably have an additional more general activation function, because expression of flo11-5 and flo11-6 is already reduced under nonstarvation conditions. The flo11-15 deletion defines a further UAS element in diploid strains that might be 3AT specific, because this deletion led to a 4.3-fold reduced expression of FLO11 in the presence of 3AT.
Three URS elements are defined by the flo11-4, flo11-7, and flo11-8 deletions. The URS sites in flo11-4 and flo11-7 do not seem to be regulated by amino acid starvation, because expression of both constructs is inducible by addition of 3AT (Table 3). In contrast, the URS element defined by the flo11-8 deletion (base pairs –1400 to –1600) couples expression of FLO11 to regulation by amino acid starvation, because its deletion led to a strong induction of FLO11-lacZ expression (7.2-fold in haploids and 77.5-fold in diploids) under nonstarvation conditions, but not in the presence of 3AT. Thus, the FLO11 promoter contains at least one 3AT-responsive URS element in segment FLO11-8 that might confer enhanced expression of FLO11 in response to amino acid starvation (Figure 6). To test, whether derepressed FLO11-lacZ expression observed for deletion of segment 8 is Gcn4p dependent, expression of flo11-8 was measured in strains lacking GCN4 and compared with expression of flo11-7 and flo11-9. We found that expression of flo11-8 was not reduced in the absence of Gcn4p, neither in the absence nor presence of 3AT. In contrast, expression of flo11-7 and flo11-9 was strongly dependent on GCN4. This further corroborates that the URS element in segment FLO11-8 is regulated by amino acid starvation and suggests that Gcn4p might be required indirectly to overcome the negative effects of this URS on FLO11 transcription.
Figure 6.
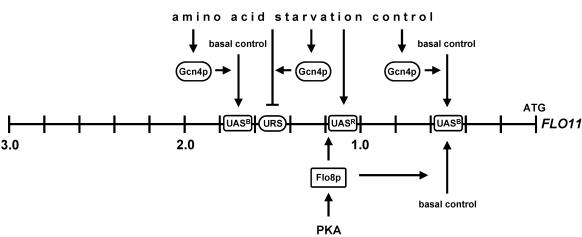
Model for regulation of FLO11 by amino acid starvation, Gcn4p, and Flo8p. Shown is the promoter of FLO11 and the regulatory elements that confer regulation by amino acid starvation (UASR and URS) or basal regulation under nonstarvation conditions (UASB). Gcn4p is postulated to control basal and amino acid starvation-regulated expression of FLO11 indirectly, whereas control of FLO11 expression by Flo8p can be indirectly or directly, as shown previously (Pan and Heitman, 2002). PKA, cAMP-dependent protein kinase.
UAS elements that mediate regulation by amino acid starvation were identified in a second approach by using a series of 14 reporter constructs containing individual 400-base pair FLO11 promoter fragments that overlap by 200 base pairs and are cloned in front of a CYC1-lacZ fusion gene (Rupp et al., 1999). This series of reporter constructs identified strong UAS elements in the segments FLO11-3/2, FLO11-6/5, FLO11-7/6, and FLO11-10/9 (Table 4 and Figure 5B). These FLO11 promoter elements increased expression of CYC1-lacZ at least fivefold compared with the reporter without any insert (Table 4). The elements present in FLO11-3/2 (base pairs –620 to –182) and FLO11-10/9 (base pairs –2020 to –1573) both confer similar activation in the absence and presence of 3AT, suggesting that they are basal UAS elements (UASB) that are not regulated by amino acid starvation (Figure 6). In contrast, activity mediated by segments FLO11-6/5 (base pairs –1220 to –779) and FLO11-7/6 (base pairs –1421 to –981) is at least 2.4-fold inducible by addition of 3AT, suggesting that these segments contain UAS elements that confer regulation by amino acid starvation (UASR). To better localize the strong basal UASB element in segment FLO11-10/9 and the regulated UASR element in segment FLO11-6/5, expression of further transcriptional reporters was measured that carried individual segments FLO11-5, FLO11-6, FLO11-9, or FLO11-10 in front of CYC1-lacZ (Table 4 and Figure 5). We found that FLO11-9, but not FLO11-10, conferred basal UAS activity, indicating that the basal UASB element is localized between base pairs –1820 to –1573 (Figure 6). Similarly, UAS activity and regulation by 3AT was conferred byFLO11-6, but not FLO11-5, indicating that the regulated UASR element is localized between base pairs –1220 to –981 (Figure 6).
Table 4.
Expression of different CYC1-lacZ reporter constructs
| Boundaries of insert
|
YNB
|
YNB + 3AT
|
3AT induction
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Inserted segment | wt | gcn4Δ | flo8Δ | wt | gcn4Δ | flo8Δ | wt | gcn4Δ | flo8Δ | |
| No insert | 7.1 | 6.9 | 7.0 | 7.7 | 7.3 | 7.3 | 1.1 | 1.0 | 1.0 | |
| 2/1 | -421 to -1 | 3.3 | 2.2 | 3.0 | 2.6 | 3.1 | 3.0 | 0.8 | 1.4 | 1.0 |
| 3/2 | -620 to -182 | 41.6 | 8.3 | 4.9 | 47.2 | 22.2 | 6.1 | 1.1 | 2.7 | 1.2 |
| 4/3 | -820 to -381 | 5.4 | 1.9 | 4.4 | 9.8 | 4.3 | 6.0 | 1.8 | 2.3 | 1.4 |
| 5/4 | -1018 to -581 | 4.5 | 4.2 | 2.6 | 7.7 | 3.9 | 2.7 | 1.7 | 0.9 | 1.0 |
| 5 | -1018 to -779 | 3.4 | 0.3 | 1.4 | 14.1 | 1.1 | 4.7 | 4.2 | 3.7 | 3.4 |
| 6/5 | -1220 to -779 | 159 | 15.2 | 3.8 | 525 | 51.1 | 25.1 | 3.3 | 3.4 | 6.6 |
| 6 | -1220 to -981 | 187 | 58.2 | 61.5 | 584 | 188 | 193 | 3.1 | 3.2 | 3.1 |
| 7/6 | -1421 to -981 | 126 | 20.8 | 8.0 | 299 | 61.7 | 27.5 | 2.4 | 2.9 | 3.5 |
| 8/7 | -1620 to -1182 | 10.8 | 4.0 | 3.9 | 13.4 | 4.8 | 4.8 | 1.2 | 1.2 | 1.2 |
| 9/8 | -1820 to -1381 | 8.6 | 5.0 | 21.0 | 18.4 | 8.4 | 13.3 | 2.1 | 1.7 | 0.6 |
| 9 | -1820 to -1573 | 101 | 18.1 | 49.8 | 128 | 23.4 | 60.1 | 1.3 | 1.3 | 1.2 |
| 10/9 | -2020 to -1573 | 194 | 32.7 | 157 | 235 | 42.9 | 186 | 1.2 | 1.3 | 1.2 |
| 10 | -2020 to -1781 | 21.4 | 2.6 | 4.0 | 27.3 | 3.1 | 7.9 | 1.3 | 1.2 | 2.0 |
| 11/10 | -2220 to -1781 | 22.4 | 1.0 | 15.1 | 36.8 | 1.2 | 25.1 | 1.6 | 1.2 | 1.7 |
| 12/11 | -2420 to -1981 | 11.3 | 3.8 | 6.8 | 10.3 | 3.8 | 6.7 | 0.9 | 1.0 | 1.0 |
| 13/12 | -2620 to -2181 | 2.1 | 1.2 | 1.5 | 2.0 | 1.8 | 2.2 | 1.0 | 1.5 | 1.5 |
| 14/13 | -2820 to -2381 | 1.7 | 1.1 | 1.4 | 1.6 | 1.9 | 1.5 | 1.0 | 1.7 | 1.0 |
| 15/14 | -2984 to -2581 | 2.1 | 1.3 | 2.0 | 1.7 | 2.1 | 2.3 | 0.8 | 1.6 | 1.2 |
Expression of indicated CYC1-lacZ reporter constructs (Rupp et al., 1999; Table 2) was assayed in the haploid strains RH2648 (wt), RH2650 (gcn4Δ) and RH2652 (flo8Δ) under nonstarvation conditions (YNB + Arg) and under amino acid starvation conditions (YNB +Arg +3AT). Segments of the FLO11 promoter inserted upstream of CYC1-lacZ in the individual constructs are indicated and correspond to the numbering shown in Figure 5. Boundaries of insertions are indicated with respect to the translational start site of FLO11 at position +1. pME2212 was used as a control plasmid carrying no insert upstream of CYCl-lacZ. Given numbers represent β-galactosidase activities in actual units nanomoles per minute per milligram. Values for 3AT induction represent the ratio of activities obtained for a given CYC1-lacZ construct in the indicated genetic background in the presence or absence of 3AT, respectively. All values are means of at least three independent measurements with a standard deviation not exceeding 15%.
FLO11 Promoter Elements Mediating Regulation by Gcn4p and Flo8p in Response to Amino Acid Starvation
Activation of FLO11-lacZ expression by 3AT is completely blocked when both Gcn4p and Flo8p are absent (Figure 4). To identify the regions of the FLO11 promoter that are under control of Gcn4p and Flo8p, the set of CYC-lacZ reporter constructs was transformed into strains deleted for GCN4 or FLO8. Deletion of GCN4 led to a more than threefold reduction in the expression of FLO11-3/2 under nonstarvation conditions, and of FLO11-5, FLO11-6/5, FLO11-6, FLO11-7/6, and FLO11-9, FLO11-10/9, FLO11-10, and FLO11-11/10 under both nonstarvation and starvation conditions. (Table 4 and Figure 5B). However, activation of FLO11-5, FLO11-6/5, FLO11-6, and FLO11-7/6 by 3AT was not significantly reduced in the absence of Gcn4p. Together, these results suggest that Gcn4p controls expression of FLO11 by affecting basal control mechanisms mediated by UASB elements, rather than by affecting UASR elements conferring regulation by amino acid starvation (Figure 6). However, regulation by Gcn4p is likely to involve further factors, because none of the Gcn4p-controlled segments of the FLO11-promoter identified herein contain a Gcn4p-binding site. In addition, none of these segments are able to bind Gcn4p protein present in yeast extracts or purified from Escherichia coli when tested in vitro (our unpublished data).
Deletion of FLO8 had significant effects on expression of FLO11-3/2, FLO11-6/5, FLO11-6, and FLO11-7/6 under both nonstarvation and amino acid starvation conditions (Table 4). These regions of the FLO11 promoter were previously identified to be under control of Flo8p (Rupp et al., 1999). Results obtained herein indicate that Flo8p might not be involved in mediating amino acid starvation signals to these elements, because FLO11-3/2 is not inducible by 3AT and because FLO11-6/5, FLO11-6, and FLO11-7/6 were still inducible by amino acid starvation in the absence of FLO8.
DISCUSSION
Cell-cell and cell-substrate adhesion are morphogenetic events that are required for several developmental processes of bakers' yeast, including mating, invasive growth, biofilm formation, and filamentation. Each of these events depends on distinct intrinsic and extrinsic signals, corresponding signaling pathways, and specific cell surface proteins. The cell surface flocculin Flo11p is required for haploid adhesive growth in response to glucose starvation and for diploid filamentous growth in response to nitrogen starvation. Herein, we found that amino acid starvation is a further nutritional signal that activates adhesive growth in a Flo11p-dependent manner and that leads to induced expression of the FLO11 gene, even in the presence of high glucose and ammonium. We have identified two distinct types of regulatory elements in the FLO11 promoter that are involved in control by amino acid starvation. One UASR element was found that is located in the region –1000 to –1200 upstream of the FLO11 open reading frame and that confers regulation by amino acid starvation (Figure 6). Regulation of this UASR, however, is independent of Gcn4p. In addition, one URS element was identified in the region –1400 to –1600 that confers repression of FLO11 under nonstarvation conditions, but not under starvation conditions, suggesting that repression might be relieved by starvation (Figure 6). Moreover, Gcn4p might be required to overcome the negative effect of this URS on FLO11 transcription. Previous studies have identified multiple cis-acting regulatory elements that confer regulation by cAMP, Flo8p, Ste12p, and Tec1p. Regulation by cAMP and Flo8p involves the region from –1000 to –1400, and Flo8p activates a further UAS element in the region from –400 to –600 (Rupp et al., 1999; Pan and Heitman, 2002). Ste12p has been reported to act on at least three elements located in regions –800 to –1200, –1600 to –1800, and –2000 to –2400, respectively, and control by Tec1p involves three elements within regions –600 to –1000 and –1600 to –2400 (Lo and Dranginis, 1998; Rupp et al., 1999). However, although some of these regulatory elements are located within the same segments that harbor the regulatory elements conferring control by amino acid starvation and Gcn4p, our study indicates that neither of the transcription factors Flo8p, Ste12p, nor Tec1p is likely to be directly involved in amino acid starvation control. Future fine analysis must reveal the exact nucleotides within the 200-base pair segment of the FLO11 promoter that confer regulation by amino acid starvation and other regulatory pathways that control FLO11 expression.
What signaling pathways are involved in mediating the amino acid starvation signal to the regulatory elements in the FLO11 promoter identified herein? We have found that Gcn2p and Gcn4p, elements of the general control system for amino acid biosynthesis, are required for amino acid starvation-induced adhesive growth and activation of FLO11. The general amino acid control system was previously unknown to regulate adhesive growth and expression of FLO11 in budding yeast (Hinnebusch and Natarajan, 2002). In the human pathogen C. albicans, amino acid starvation and Gcn4p have been found to affect hyphal morphogenesis (Tripathi et al., 2002). However, whether cell-substrate adhesion and expression of specific cell-surface proteins is induced by amino acid starvation and depends on Gcn2p and Gcn4p has not been shown in C. albicans. Several observations suggest that expression of FLO11 might not involve direct binding of Gcn4p to the FLO11 promoter and that an increase in protein levels of Gcn4p per se is not sufficient for enhanced FLO11 transcription. 1) Sequence analysis of the FLO11 promoter does not predict any Gcn4p-responsive element sites. 2) Gcn4p protein does not bind to any region of the FLO11 promoter when tested in vitro (our unpublished data). 3) High-level expression of Gcn4p in nonstarved cells is not sufficient to induce enhanced expression of FLO11. However, a scenario in which Gcn4p directly binds to the FLO11-promoter in combination with other transcriptional regulators cannot be ruled out by our data. We suggest two roles for Gcn4p in regulating expression of FLO11 (Figure 6). First, Gcn4p regulates expression of FLO11 by affecting activity of the two basal UASB elements present in the regions –400 to –600 and –1600 to –1800. This conclusion is based on the finding that FLO11 expression drops significantly in the absence of Gcn4p and that both UASB elements require Gcn4p to mediate efficient activation. Gcn4p seems to control these UASB elements indirectly, because neither of them contains a consensus Gcn4p-binding site. Again, Gcn4p might directly regulate these elements in concert with other transcriptional factors by contacting yet unknown DNA sequence elements. The second role of Gcn4p suggested by our results is control of a pathway that confers relieve of URS-mediated repression of FLO11 transcription in response to amino acid starvation. This conclusion is based on the finding that deletion of the URS located in the region –1400 to –1600 causes derepressed expression of FLO11 independent of Gcn4p. The exact DNA sequence elements conferring repression and the pathway required for relieve of repression remain to be determined by future investigations.
We have identified two central elements of the cAMP pathway, Tpk2p and Flo8p, to be required for amino acid starvation-induced adhesive growth and activation of FLO11. Previous studies have shown that the protein kinase Tpk2p together with the transcriptional regulators Flo8p and Sfl1p confer regulation of FLO11 in response to cAMP (Robertson and Fink, 1998; Pan and Heitman, 1999; Rupp et al., 1999; Pan and Heitman, 2002). Flo8p and Sfl1p are direct molecular targets of Tpk2p that antagonistically regulate expression of FLO11 via a common promoter element located within base pairs –1400 to –1150 (Pan and Heitman, 2002). Herein, we found that Tpk2p and Flo8p are required for 3AT-induced expression of FLO11, which involves the UASR element located within base pairs –1200 to –1000, and the 3AT responsive URS element located within base pairs –1600 to –1400 (Figure 6). Flo8p is not likely to be involved in directly mediating the amino acid starvation signal to the FLO11 promoter, because none of the 3AT-responsive elements in the FLO11 promoter requires Flo8p for activation by amino acid starvation. Our data rather suggest that Flo8p is required for basal expression of FLO11 and that absence of Flo8p cannot be compensated by amino acid starvation. Absence of Tpk2p is likely to cause repression of FLO11 by efficient binding of Sfl1p to segment –1400 to –1150, a transcriptional block that cannot be relieved by amino acid starvation. Our study also disfavors involvement of the filamentous/invasive MAPK cascade in mediation of the amino acid starvation signal to FLO11, because neither Ste12p nor Tec1p were found to be required for activation by 3AT.
We have uncovered amino acid starvation as a nutritional signal that in S. cerevisiae efficiently activates adhesive growth and expression of FLO11, even when high amounts of preferred carbon and nitrogen sources are available. These growth conditions reflect the nutritional situation of yeast cells that grow on fruits, a natural habitat of S. cerevisiae. Fruits are rich in carbon sources such as saccharose or glucose and contain a variety of nitrogen sources at ample concentrations (Bisson, 1991). The content of different amino acids, however, is highly unbalanced in fruits. In grapes, an important natural substrate for S. cerevisiae, proline and arginine are the predominant amino acids, and their concentration often exceeds that found for histidine by 10–100 times (Huang and Ough, 1989). External amino acid imbalance is one of the signals that activate the general control system in S. cerevisiae (Niederberger et al., 1981). In conclusion, our study suggests that starvation for amino acids rather than a general lack of carbon or nitrogen sources might be the nutritional signal that activates cell-cell and cell-surface adhesion of yeast living on the natural habitat.
Acknowledgments
We are grateful to Maria Meyer for excellent technical assistance during the course of this work. We thank Anne Dranginis, Gerald R. Fink, Haoping Liu, and Steffen Rupp for generously providing plasmids and yeast strains. G.H.B. thanks Mark I. Cockett (Bristol-Myers Squibb Functional Genomics Institute, Hopewell Campus, Pennington NY) for the possibility to spend sabbatical leave. This work was supported by grants from the Deutsche Forschungsgemeinschaft, the Volkswagenstiftung, and Fonds der Chemischen Industrie.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E03–01–0042. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E03-01-0042.
References
- Albrecht, G., Mösch, H.-U., Hoffmann, B., Reusser, U., and Braus, G.H. (1998). Monitoring the Gcn4 protein-mediated response in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 273, 12696–12702. [DOI] [PubMed] [Google Scholar]
- Ausubel, F.M., Brent, R., Kingston, R.E., Moore, D.D., Seidman, J.G., Smith, J.A., and Struhl, K. (1993). Current Protocols in Molecular Biology. New York: Greene Publishing Associates and Wiley Interscience.
- Banuett, F. (1998). Signalling in the yeasts: an informational cascade with links to the filamentous fungi. Microbiol. Mol. Biol. Rev. 62, 249–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisson, L.F. (1991). Influence of nitrogen on yeast and fermentation of grapes. In: Proceedings of the International Symposium on Nitrogen in Grapes and Wine, Seattle, WA: American Society of Enology and Viticulture.
- Bradford, M. (1976). A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254. [DOI] [PubMed] [Google Scholar]
- Cappellaro, C., Baldermann, C., Rachel, R., and Tanner, W. (1994). Mating type-specific cell-cell recognition of Saccharomyces cerevisiae: cell wall attachment and active sites of a- and alpha-agglutinin. EMBO J. 13, 4737–4744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross, F.R., and Tinkelenberg, A.H. (1991). A potential positive feedback loop controlling CLN1 and CLN2 gene expression at the start of the yeast cell cycle. Cell 65, 875–883. [DOI] [PubMed] [Google Scholar]
- Cullen, P.J., and Sprague, G.F., Jr. (2000). Glucose depletion causes haploid invasive growth in yeast. Proc. Natl. Acad. Sci. USA 97, 13619–13624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz, R.D., and Sugino, A. (1988). New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene 74, 527–534. [DOI] [PubMed] [Google Scholar]
- Gimeno, C.J., Ljungdahl, P.O., Styles, C.A., and Fink, G.R. (1992). Unipolar cell divisions in the yeast S. cerevisiae lead to filamentous growth: regulation by starvation and RAS. Cell 68, 1077–1090. [DOI] [PubMed] [Google Scholar]
- Grundmann, O., Mösch, H.-U., and Braus, G.H. (2001). Repression of GCN4 mRNA translation by nitrogen starvation in Saccharomyces cerevisiae. J. Biol. Chem. 276, 25661–25671. [DOI] [PubMed] [Google Scholar]
- Guarente, L., and Ptashne, M. (1981). Fusion of Escherichia coli lacZ to the cytochrome c gene of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 78, 2199–2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, B., Styles, C.A., Feng, Q., and Fink, G.R. (2000). A Saccharomyces gene family involved in invasive growth, cell-cell adhesion, and mating. Proc. Natl. Acad. Sci. USA 97, 12158–12163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guthrie, C., and Fink, G.R. (1991). Guide to Yeast Genetics and Molecular Biology. San Diego, CA: Academic Press.
- Hilton, J.L., Kearney, P.C., and Ames, B.N. (1965). Mode of action of the herbicide, 3-amino-1,2,4-triazole(amitrole): inhibition of an enzyme of histidine biosynthesis. Arch. Biochem. Biophys. 112, 544–547. [DOI] [PubMed] [Google Scholar]
- Hinnebusch, A.G. (1986). The general control of amino acid biosynthetic genes in the yeast Saccharomyces cerevisiae. Crit. Rev. Biochem. 21, 277–317. [DOI] [PubMed] [Google Scholar]
- Hinnebusch, A.G. (1992). General and pathway-specific regulatory mechanisms controlling the synthesis of amino acid biosynthetic enzymes in Saccharomyces cerevisiae. In: The Molecular and Cellular Biology of the Yeast Saccharomyces cerevisiae: Gene Expression, ed. J.R. Broach, E.W. Jones, and J.R. Pringle, Cold Spring Harbor, NY: Cold Spring Harbor Laboratory, 319–414.
- Hinnebusch, A.G. (1997). Translational regulation of yeast GCN4. A window on factors that control initiator-tRNA binding to the ribosome. J. Biol. Chem. 272, 21661–21664. [DOI] [PubMed] [Google Scholar]
- Hinnebusch, A.G., and Natarajan, K. (2002). Gcn4p, a master regulator of gene expression, is controlled at multiple levels by diverse signals of starvation and stress. Eukaryote Cell 1, 22–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hope, I.A., and Struhl, K. (1985). GCN4 protein, synthesized in vitro, binds HIS3 regulatory sequences: implications for general control of amino acid biosynthetic genes in yeast. Cell 43, 177–188. [DOI] [PubMed] [Google Scholar]
- Huang, Z., and Ough, C.S. (1989). Effect of vineyard locations, varieties, and rootstocks on the juice amino acid composition of several cultivars. Am. J. Enol. Vitic. 40, 135–139. [Google Scholar]
- Ito, H., Fukuda, Y., Murata, K., and Kimura, A. (1983). Transformation of intact yeast cells treated with alkali cations. J. Bacteriol. 153, 163–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia, M.H., Larossa, R.A., Lee, J.M., Rafalski, A., Derose, E., Gonye, G., and Xue, Z. (2000). Global expression profiling of yeast treated with an inhibitor of amino acid biosynthesis, sulfometuron methyl. Physiol. Genomics 3, 83–92. [DOI] [PubMed] [Google Scholar]
- Köhler, T., Wesche, S., Taheri, N., Braus, G.H., and Mösch, H.-U. (2002). Dual role of the Saccharomyces cerevisiae TEA/ATTS family transcription factor Tec1p in regulation of gene expression and cellular development. Eukaryote Cell 1, 673–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuchin, S., Vyas, V.K., and Carlson, M. (2002). Snf1 protein kinase and the repressors Nrg1 and Nrg2 regulate FLO11, haploid invasive growth, and diploid pseudohyphal differentiation. Mol. Cell. Biol. 22, 3994–4000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengeler, K.B., Davidson, R.C., D'Souza, C., Harashima, T., Shen, W.C., Wang, P., Pan, X., Waugh, M., and Heitman, J. (2000). Signal transduction cascades regulating fungal development and virulence. Microbiol. Mol. Biol. Rev. 64, 746–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, H., Styles, C.A., and Fink, G.R. (1993). Elements of the yeast pheromone response pathway required for filamentous growth of diploids. Science 262, 1741–1744. [DOI] [PubMed] [Google Scholar]
- Liu, H., Styles, C.A., and Fink, G.R. (1996). Saccharomyces cerevisiae S288C has a mutation in FLO8, a gene required for filamentous growth. Genetics 144, 967–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo, W.S., and Dranginis, A.M. (1996). FLO11, a yeast gene related to the STA genes, encodes a novel cell surface flocculin. J. Bacteriol. 178, 7144–7151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo, W.S., and Dranginis, A.M. (1998). The cell surface flocculin Flo11 is required for pseudohyphae formation and invasion by Saccharomyces cerevisiae. Mol. Biol. Cell 9, 161–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meussdoerffer, F., and Fink, G.R. (1983). Structure and expression of two aminoacyl-tRNA synthetase genes from Saccharomyces cerevisiae. J. Biol. Chem. 258, 6293–6299. [PubMed] [Google Scholar]
- Mirande, M., and Waller, J.P. (1988). The yeast lysyl-tRNA synthetase gene. Evidence for general amino acid control of its expression and domain structure of the encoded protein. J. Biol. Chem. 263, 18443–18451. [PubMed] [Google Scholar]
- Mösch, H.-U., and Fink, G.R. (1997). Dissection of filamentous growth by transposon mutagenesis in Saccharomyces cerevisiae. Genetics 145, 671–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mösch, H.-U., Kübler, E., Krappmann, S., Fink, G.R., and Braus, G.H. (1999). Crosstalk between the Ras2p-controlled mitogen-activated protein kinase and cAMP pathways during invasive growth of Saccharomyces cerevisiae. Mol. Biol. Cell 10, 1325–1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mösch, H.-U., Scheier, B., Lahti, R., Mäntsälä, P., and Braus, G.H. (1991). Transcriptional activation of yeast nucleotide biosynthetic gene ADE4 by GCN4. J. Biol. Chem. 266, 20453–20456. [PubMed] [Google Scholar]
- Mueller, P.P., and Hinnebusch, A.G. (1986). Multiple upstream AUG codons mediate translational control of. GCN4. Cell 45, 201–207. [DOI] [PubMed] [Google Scholar]
- Natarajan, K., Meyer, M.R., Jackson, B.M., Slade, D., Roberts, C., Hinnebusch, A.G., and Marton, M.J. (2001). Transcriptional profiling shows that Gcn4p is a master regulator of gene expression during amino acid starvation in yeast. Mol. Cell. Biol. 21, 4347–4368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niederberger, P., Miozzari, G., and Hütter, R. (1981). Biological role of the general control of amino acid biosynthesis in Saccharomyces cerevisiae. Mol. Cell. Biol. 1, 584–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliphant, A.R., Brandl, C.J., and Struhl, K. (1989). Defining the sequence specificity of DNA-binding proteins by selecting binding sites from random-sequence oligonucleotides: analysis of yeast GCN4 protein. Mol. Cell. Biol. 9, 2944–2949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan, X., and Heitman, J. (1999). Cyclic AMP-dependent protein kinase regulates pseudohyphal differentiation in Saccharomyces cerevisiae. Mol. Cell. Biol. 19, 4874–4887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan, X., and Heitman, J. (2002). Protein kinase A operates a molecular switch that governs yeast pseudohyphal differentiation. Mol. Cell. Biol. 22, 3981–3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds, T.B., and Fink, G.R. (2001). Bakers'yeast, a model for fungal biofilm formation. Science 291, 878–881. [DOI] [PubMed] [Google Scholar]
- Roberts, R.L., and Fink, G.R. (1994). Elements of a single MAP kinase cascade in Saccharomyces cerevisiae mediate two developmental programs in the same cell type: mating and invasive growth. Genes Dev. 8, 2974–2985. [DOI] [PubMed] [Google Scholar]
- Robertson, L.S., and Fink, G.R. (1998). The three yeast A kinases have specific signaling functions in pseudohyphal growth. Proc. Natl. Acad. Sci. USA 95, 13783–13787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose, M., and Botstein, D. (1983). Construction and use of gene fusions to lacZ (β-galactosidase) that are expressed in yeast. Methods Enzymol. 101, 167–180. [DOI] [PubMed] [Google Scholar]
- Roy, A., Lu, C.F., Marykwas, D.L., Lipke, P.N., and Kurjan, J. (1991). The AGA1 product is involved in cell surface attachment of the Saccharomyces cerevisiae cell adhesion glycoprotein a-agglutinin. Mol. Cell. Biol. 11, 4196–4206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupp, S., Summers, E., Lo, H.J., Madhani, H., and Fink, G.R. (1999). MAP kinase and cAMP filamentation signaling pathways converge on the unusually large promoter of the yeast FLO11 gene. EMBO J. 18, 1257–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schürch, A., Miozzari, J., and Hütter, R. (1974). Regulation of tryptophan biosynthesis in Saccharomyces cerevisiae: mode of action of 5-methyl-tryptophan and 5-methyl-tryptophan-sensitive mutants. J. Bacteriol. 117, 1131–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi, G., Wiltshire, C., Macaskill, S., Tournu, H., Budge, S., and Brown, A.J. (2002). Gcn4 co-ordinates morphogenetic and metabolic responses to amino acid starvation in Candida albicans. EMBO J. 21, 5448–5456. [DOI] [PMC free article] [PubMed] [Google Scholar]