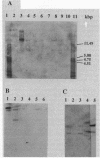
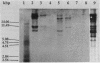
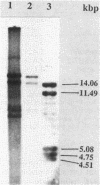
Abstract
The conjugative streptococcal transposon Tn916 was found to transfer naturally between a variety of gram-positive and gram-negative eubacteria. Enterococcus faecalis hosting the transposon could serve as a donor for Alcaligenes eutrophus, Citrobacter freundii, and Escherichia coli at frequencies of 10(-6) to 10(-8). No transfer was observed with several phototrophic species. Mating of an E. coli strain carrying Tn916 yielded transconjugants with Bacillus subtilis, Clostridium acetobutylicum, Enterococcus faecalis, and Streptococcus lactis subsp. diacetylactis at frequencies of 10(-4) to 10(-6). Acetobacterium woodii was the only gram-positive organism tested that did not accept the transposon from a gram-negative donor. The results prove the ability of conjugative transposable elements such as Tn916 for natural cross-species gene transfer, thus potentially contributing to bacterial evolution.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bannister J. V., Parker M. W. The presence of a copper/zinc superoxide dismutase in the bacterium Photobacterium leiognathi: a likely case of gene transfer from eukaryotes to prokaryotes. Proc Natl Acad Sci U S A. 1985 Jan;82(1):149–152. doi: 10.1073/pnas.82.1.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer H. W., Roulland-Dussoix D. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J Mol Biol. 1969 May 14;41(3):459–472. doi: 10.1016/0022-2836(69)90288-5. [DOI] [PubMed] [Google Scholar]
- Brisson-Noël A., Arthur M., Courvalin P. Evidence for natural gene transfer from gram-positive cocci to Escherichia coli. J Bacteriol. 1988 Apr;170(4):1739–1745. doi: 10.1128/jb.170.4.1739-1745.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang A. C., Cohen S. N. Genome construction between bacterial species in vitro: replication and expression of Staphylococcus plasmid genes in Escherichia coli. Proc Natl Acad Sci U S A. 1974 Apr;71(4):1030–1034. doi: 10.1073/pnas.71.4.1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie P. J., Dunny G. M. Identification of regions of the Streptococcus faecalis plasmid pCF-10 that encode antibiotic resistance and pheromone response functions. Plasmid. 1986 May;15(3):230–241. doi: 10.1016/0147-619x(86)90041-7. [DOI] [PubMed] [Google Scholar]
- Christie P. J., Korman R. Z., Zahler S. A., Adsit J. C., Dunny G. M. Two conjugation systems associated with Streptococcus faecalis plasmid pCF10: identification of a conjugative transposon that transfers between S. faecalis and Bacillus subtilis. J Bacteriol. 1987 Jun;169(6):2529–2536. doi: 10.1128/jb.169.6.2529-2536.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clewell D. B., Flannagan S. E., Ike Y., Jones J. M., Gawron-Burke C. Sequence analysis of termini of conjugative transposon Tn916. J Bacteriol. 1988 Jul;170(7):3046–3052. doi: 10.1128/jb.170.7.3046-3052.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clewell D. B., Gawron-Burke C. Conjugative transposons and the dissemination of antibiotic resistance in streptococci. Annu Rev Microbiol. 1986;40:635–659. doi: 10.1146/annurev.mi.40.100186.003223. [DOI] [PubMed] [Google Scholar]
- Courvalin P., Carlier C. Tn1545: a conjugative shuttle transposon. Mol Gen Genet. 1987 Feb;206(2):259–264. doi: 10.1007/BF00333582. [DOI] [PubMed] [Google Scholar]
- Frank-Kamenetskii M. DNA structure: physicists retreat again. Nature. 1987 Jul 9;328(6126):108–108. doi: 10.1038/328108a0. [DOI] [PubMed] [Google Scholar]
- Gawron-Burke C., Clewell D. B. Regeneration of insertionally inactivated streptococcal DNA fragments after excision of transposon Tn916 in Escherichia coli: strategy for targeting and cloning of genes from gram-positive bacteria. J Bacteriol. 1984 Jul;159(1):214–221. doi: 10.1128/jb.159.1.214-221.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinemann J. A., Sprague G. F., Jr Bacterial conjugative plasmids mobilize DNA transfer between bacteria and yeast. Nature. 1989 Jul 20;340(6230):205–209. doi: 10.1038/340205a0. [DOI] [PubMed] [Google Scholar]
- Horodniceanu T., Bougueleret L., El-Solh N., Bouanchaud D. H., Chabbert Y. A. Conjugative R plasmids in Streptococcus agalactiae (group B). Plasmid. 1979 Apr;2(2):197–206. doi: 10.1016/0147-619x(79)90038-6. [DOI] [PubMed] [Google Scholar]
- Hächler H., Kayser F. H., Berger-Bächi B. Homology of a transferable tetracycline resistance determinant of Clostridium difficile with Streptococcus (Enterococcus) faecalis transposon Tn916. Antimicrob Agents Chemother. 1987 Jul;31(7):1033–1038. doi: 10.1128/aac.31.7.1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreft J., Bernhard K., Goebel W. Recombinant plasmids capable to replication in B. subtilis and E. coli. Mol Gen Genet. 1978 Jun 1;162(1):59–67. doi: 10.1007/BF00333851. [DOI] [PubMed] [Google Scholar]
- Lin Y. L., Blaschek H. P. Transformation of Heat-Treated Clostridium acetobutylicum Protoplasts with pUB110 Plasmid DNA. Appl Environ Microbiol. 1984 Oct;48(4):737–742. doi: 10.1128/aem.48.4.737-742.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazodier P., Petter R., Thompson C. Intergeneric conjugation between Escherichia coli and Streptomyces species. J Bacteriol. 1989 Jun;171(6):3583–3585. doi: 10.1128/jb.171.6.3583-3585.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray B. E., An F. Y., Clewell D. B. Plasmids and pheromone response of the beta-lactamase producer Streptococcus (Enterococcus) faecalis HH22. Antimicrob Agents Chemother. 1988 Apr;32(4):547–551. doi: 10.1128/aac.32.4.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien R. W., Morris J. G. Oxygen and the growth and metabolism of Clostridium acetobutylicum. J Gen Microbiol. 1971 Nov;68(3):307–318. doi: 10.1099/00221287-68-3-307. [DOI] [PubMed] [Google Scholar]
- Roberts M. C., Koutsky L. A., Holmes K. K., LeBlanc D. J., Kenny G. E. Tetracycline-resistant Mycoplasma hominis strains contain streptococcal tetM sequences. Antimicrob Agents Chemother. 1985 Jul;28(1):141–143. doi: 10.1128/aac.28.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouch D. A., Skurray R. A. IS257 from Staphylococcus aureus: member of an insertion sequence superfamily prevalent among gram-positive and gram-negative bacteria. Gene. 1989;76(2):195–205. doi: 10.1016/0378-1119(89)90160-1. [DOI] [PubMed] [Google Scholar]
- Schäfer A., Kalinowski J., Simon R., Seep-Feldhaus A. H., Pühler A. High-frequency conjugal plasmid transfer from gram-negative Escherichia coli to various gram-positive coryneform bacteria. J Bacteriol. 1990 Mar;172(3):1663–1666. doi: 10.1128/jb.172.3.1663-1666.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott J. R., Kirchman P. A., Caparon M. G. An intermediate in transposition of the conjugative transposon Tn916. Proc Natl Acad Sci U S A. 1988 Jul;85(13):4809–4813. doi: 10.1073/pnas.85.13.4809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Strätz M., Gottschalk G., Dürre P. Transfer and expression of the tetracycline resistance transposon Tn925 in Acetobacterium woodii. FEMS Microbiol Lett. 1990 Mar 1;56(1-2):171–176. doi: 10.1111/j.1574-6968.1990.tb04144.x. [DOI] [PubMed] [Google Scholar]
- Trieu-Cuot P., Gerbaud G., Lambert T., Courvalin P. In vivo transfer of genetic information between gram-positive and gram-negative bacteria. EMBO J. 1985 Dec 16;4(13A):3583–3587. doi: 10.1002/j.1460-2075.1985.tb04120.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese C. R. Bacterial evolution. Microbiol Rev. 1987 Jun;51(2):221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolin E. A., Wolfe R. S., Wolin M. J. Viologen dye inhibition of methane formation by Methanobacillus omelianskii. J Bacteriol. 1964 May;87(5):993–998. doi: 10.1128/jb.87.5.993-998.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]