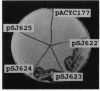
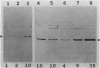
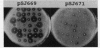
Abstract
The lipA gene encoding an extracellular lipase from Pseudomonas cepacia was cloned and sequenced. Downstream from the lipase gene an open reading frame was identified, and the corresponding gene was named limA. lipA was well expressed only in the presence of limA. limA exerts its effect both in cis and in trans and therefore produces a diffusible gene product, presumably a protein of 344 amino acids. Replacement of the lipA expression signals (promoter, ribosome-binding site, and signal peptide-coding sequences) by heterologous signals from gram-positive bacteria still resulted in limA-dependent lipA expression in Escherichia coli, Bacillus subtilis, and Streptomyces lividans.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Antonian E. Recent advances in the purification, characterization and structure determination of lipases. Lipids. 1988 Dec;23(12):1101–1106. doi: 10.1007/BF02535273. [DOI] [PubMed] [Google Scholar]
- Aoyama S., Yoshida N., Inouye S. Cloning, sequencing and expression of the lipase gene from Pseudomonas fragi IFO-12049 in E. coli. FEBS Lett. 1988 Dec 19;242(1):36–40. doi: 10.1016/0014-5793(88)80980-3. [DOI] [PubMed] [Google Scholar]
- Bibb M. J., Findlay P. R., Johnson M. W. The relationship between base composition and codon usage in bacterial genes and its use for the simple and reliable identification of protein-coding sequences. Gene. 1984 Oct;30(1-3):157–166. doi: 10.1016/0378-1119(84)90116-1. [DOI] [PubMed] [Google Scholar]
- Buttner M J, Fearnley I M, Bibb M J. The agarase gene (dagA) of Streptomyces coelicolor A3(2): nucleotide sequence and transcriptional analysis. Mol Gen Genet. 1987 Aug;209(1):101–109. doi: 10.1007/BF00329843. [DOI] [PubMed] [Google Scholar]
- Chang A. C., Cohen S. N. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol. 1978 Jun;134(3):1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diderichsen B., Wedsted U., Hedegaard L., Jensen B. R., Sjøholm C. Cloning of aldB, which encodes alpha-acetolactate decarboxylase, an exoenzyme from Bacillus brevis. J Bacteriol. 1990 Aug;172(8):4315–4321. doi: 10.1128/jb.172.8.4315-4321.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dretzen G., Bellard M., Sassone-Corsi P., Chambon P. A reliable method for the recovery of DNA fragments from agarose and acrylamide gels. Anal Biochem. 1981 Apr;112(2):295–298. doi: 10.1016/0003-2697(81)90296-7. [DOI] [PubMed] [Google Scholar]
- Frischauf A. M., Lehrach H., Poustka A., Murray N. Lambda replacement vectors carrying polylinker sequences. J Mol Biol. 1983 Nov 15;170(4):827–842. doi: 10.1016/s0022-2836(83)80190-9. [DOI] [PubMed] [Google Scholar]
- Gergen J. P., Stern R. H., Wensink P. C. Filter replicas and permanent collections of recombinant DNA plasmids. Nucleic Acids Res. 1979 Dec 20;7(8):2115–2136. doi: 10.1093/nar/7.8.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gryczan T. J., Contente S., Dubnau D. Characterization of Staphylococcus aureus plasmids introduced by transformation into Bacillus subtilis. J Bacteriol. 1978 Apr;134(1):318–329. doi: 10.1128/jb.134.1.318-329.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Götz F., Popp F., Korn E., Schleifer K. H. Complete nucleotide sequence of the lipase gene from Staphylococcus hyicus cloned in Staphylococcus carnosus. Nucleic Acids Res. 1985 Aug 26;13(16):5895–5906. doi: 10.1093/nar/13.16.5895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattori M., Sakaki Y. Dideoxy sequencing method using denatured plasmid templates. Anal Biochem. 1986 Feb 1;152(2):232–238. doi: 10.1016/0003-2697(86)90403-3. [DOI] [PubMed] [Google Scholar]
- Henikoff S. Unidirectional digestion with exonuclease III creates targeted breakpoints for DNA sequencing. Gene. 1984 Jun;28(3):351–359. doi: 10.1016/0378-1119(84)90153-7. [DOI] [PubMed] [Google Scholar]
- Kieser T. Factors affecting the isolation of CCC DNA from Streptomyces lividans and Escherichia coli. Plasmid. 1984 Jul;12(1):19–36. doi: 10.1016/0147-619x(84)90063-5. [DOI] [PubMed] [Google Scholar]
- Kieser T., Melton R. E. Plasmid pIJ699, a multi-copy positive-selection vector for Streptomyces. Gene. 1988 May 15;65(1):83–91. doi: 10.1016/0378-1119(88)90419-2. [DOI] [PubMed] [Google Scholar]
- Kugimiya W., Otani Y., Hashimoto Y., Takagi Y. Molecular cloning and nucleotide sequence of the lipase gene from Pseudomonas fragi. Biochem Biophys Res Commun. 1986 Nov 26;141(1):185–190. doi: 10.1016/s0006-291x(86)80352-7. [DOI] [PubMed] [Google Scholar]
- Lee C. Y., Iandolo J. J. Lysogenic conversion of staphylococcal lipase is caused by insertion of the bacteriophage L54a genome into the lipase structural gene. J Bacteriol. 1986 May;166(2):385–391. doi: 10.1128/jb.166.2.385-391.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C. Y., Iandolo J. J. Mechanism of bacteriophage conversion of lipase activity in Staphylococcus aureus. J Bacteriol. 1985 Oct;164(1):288–293. doi: 10.1128/jb.164.1.288-293.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel M., Higa A. Calcium-dependent bacteriophage DNA infection. J Mol Biol. 1970 Oct 14;53(1):159–162. doi: 10.1016/0022-2836(70)90051-3. [DOI] [PubMed] [Google Scholar]
- McClure W. R. Mechanism and control of transcription initiation in prokaryotes. Annu Rev Biochem. 1985;54:171–204. doi: 10.1146/annurev.bi.54.070185.001131. [DOI] [PubMed] [Google Scholar]
- Oropeza-Wekerle R. L., Müller E., Kern P., Meyermann R., Goebel W. Synthesis, inactivation, and localization of extracellular and intracellular Escherichia coli hemolysins. J Bacteriol. 1989 May;171(5):2783–2788. doi: 10.1128/jb.171.5.2783-2788.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shine J., Dalgarno L. The 3'-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc Natl Acad Sci U S A. 1974 Apr;71(4):1342–1346. doi: 10.1073/pnas.71.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens M. A., Ortlepp S. A., Ollington J. F., McConnell D. J. Nucleotide sequence of the 5' region of the Bacillus licheniformis alpha-amylase gene: comparison with the B. amyloliquefaciens gene. J Bacteriol. 1984 Apr;158(1):369–372. doi: 10.1128/jb.158.1.369-372.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strathdee C. A., Lo R. Y. Cloning, nucleotide sequence, and characterization of genes encoding the secretion function of the Pasteurella haemolytica leukotoxin determinant. J Bacteriol. 1989 Feb;171(2):916–928. doi: 10.1128/jb.171.2.916-928.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner W., Vogel M., Goebel W. Transport of hemolysin across the outer membrane of Escherichia coli requires two functions. J Bacteriol. 1983 Apr;154(1):200–210. doi: 10.1128/jb.154.1.200-210.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West S. E., Iglewski B. H. Codon usage in Pseudomonas aeruginosa. Nucleic Acids Res. 1988 Oct 11;16(19):9323–9335. doi: 10.1093/nar/16.19.9323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohlfarth S., Winkler U. K. Chromosomal mapping and cloning of the lipase gene of Pseudomonas aeruginosa. J Gen Microbiol. 1988 Feb;134(2):433–440. doi: 10.1099/00221287-134-2-433. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- Yasbin R. E., Wilson G. A., Young F. E. Transformation and transfection in lysogenic strains of Bacillus subtilis: evidence for selective induction of prophage in competent cells. J Bacteriol. 1975 Jan;121(1):296–304. doi: 10.1128/jb.121.1.296-304.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- d'Enfert C., Ryter A., Pugsley A. P. Cloning and expression in Escherichia coli of the Klebsiella pneumoniae genes for production, surface localization and secretion of the lipoprotein pullulanase. EMBO J. 1987 Nov;6(11):3531–3538. doi: 10.1002/j.1460-2075.1987.tb02679.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Heijne G. A new method for predicting signal sequence cleavage sites. Nucleic Acids Res. 1986 Jun 11;14(11):4683–4690. doi: 10.1093/nar/14.11.4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Heijne G. Signal sequences. The limits of variation. J Mol Biol. 1985 Jul 5;184(1):99–105. doi: 10.1016/0022-2836(85)90046-4. [DOI] [PubMed] [Google Scholar]