Abstract
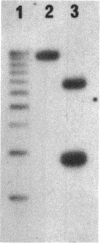
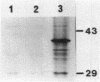
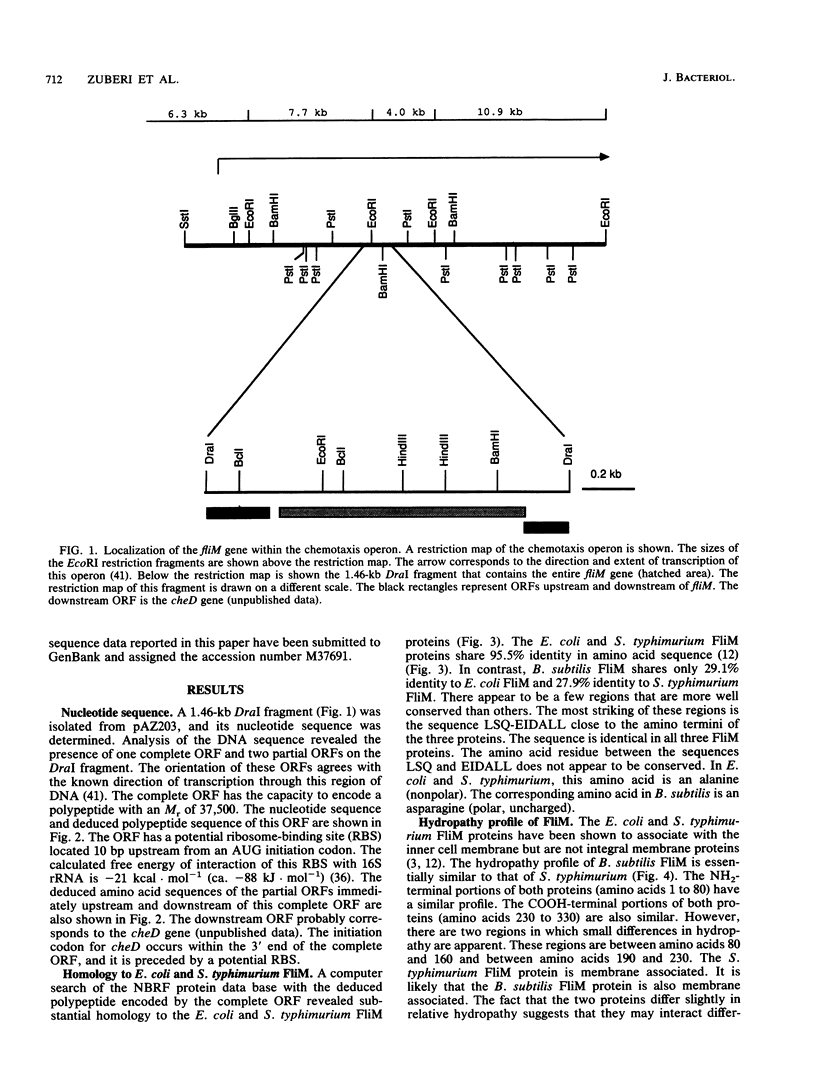
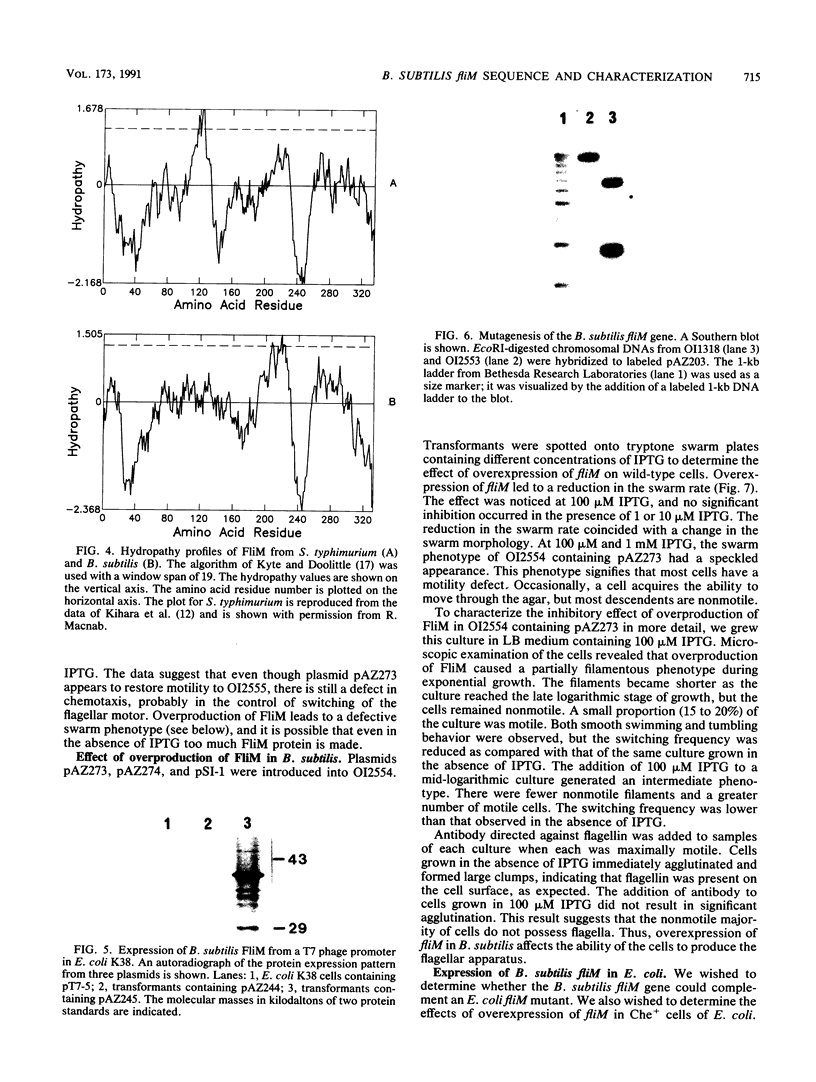
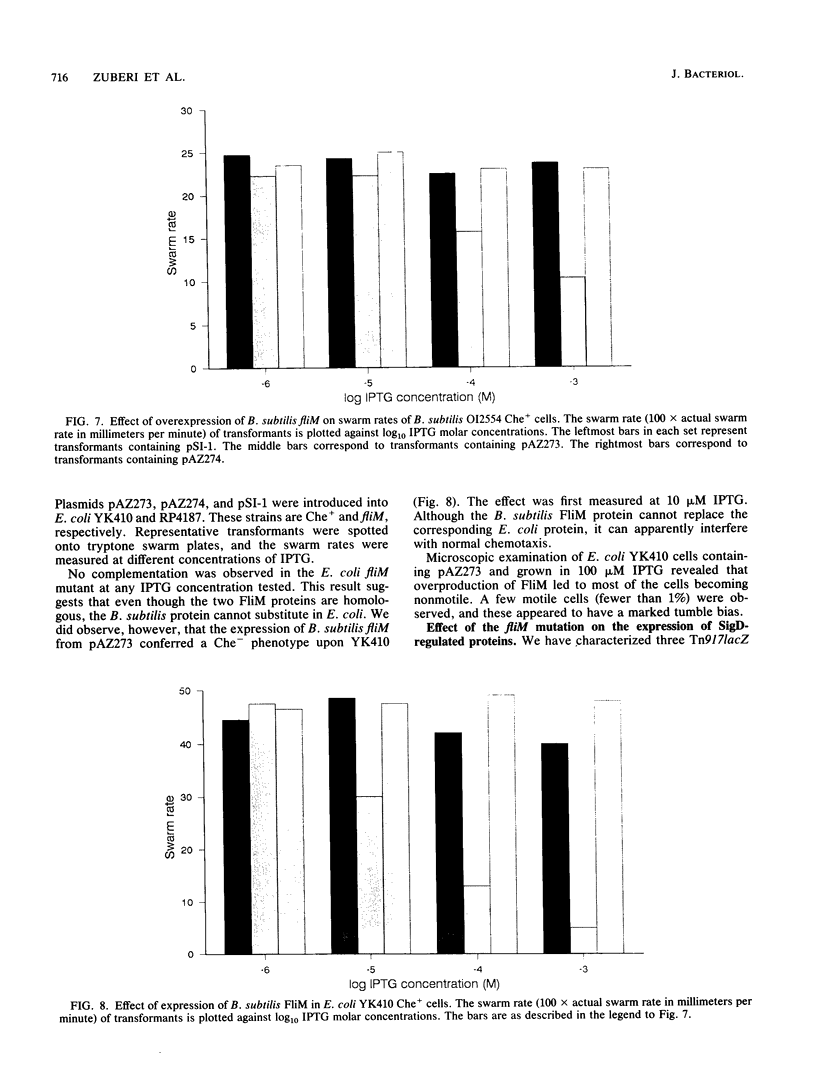
The nucleotide sequence of the Bacillus subtilis fliM gene has been determined. This gene encodes a 38-kDa protein that is homologous to the FliM flagellar switch proteins of Escherichia coli and Salmonella typhimurium. Expression of this gene in Che+ cells of E. coli and B. subtilis interferes with normal chemotaxis. The nature of the chemotaxis defect is dependent upon the host used. In B. subtilis, overproduction of FliM generates mostly nonmotile cells. Those cells that are motile switch less frequently. Expression of B. subtilis FliM in E. coli also generates nonmotile cells. However, those cells that are motile have a tumble bias. The B. subtilis fliM gene cannot complement an E. coli fliM mutant. A frameshift mutation was constructed in the fliM gene, and the mutation was transferred onto the B. subtilis chromosome. The mutant has a Fla- phenotype. This phenotype is consistent with the hypothesis that the FliM protein encodes a component of the flagellar switch in B. subtilis. Additional characterization of the fliM mutant suggests that the hag and mot loci are not expressed. These loci are regulated by the SigD form of RNA polymerase. We also did not observe any methyl-accepting chemotaxis proteins in an in vivo methylation experiment. The expression of these proteins is also dependent upon SigD. It is possible that a functional basal body-hook complex may be required for the expression of SigD-regulated chemotaxis and motility genes.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Biggin M. D., Gibson T. J., Hong G. F. Buffer gradient gels and 35S label as an aid to rapid DNA sequence determination. Proc Natl Acad Sci U S A. 1983 Jul;80(13):3963–3965. doi: 10.1073/pnas.80.13.3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourret R. B., Hess J. F., Simon M. I. Conserved aspartate residues and phosphorylation in signal transduction by the chemotaxis protein CheY. Proc Natl Acad Sci U S A. 1990 Jan;87(1):41–45. doi: 10.1073/pnas.87.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clegg D. O., Koshland D. E., Jr Identification of a bacterial sensing protein and effects of its elevated expression. J Bacteriol. 1985 Apr;162(1):398–405. doi: 10.1128/jb.162.1.398-405.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driks A., DeRosier D. J. Additional structures associated with bacterial flagellar basal body. J Mol Biol. 1990 Feb 20;211(4):669–672. doi: 10.1016/0022-2836(90)90063-R. [DOI] [PubMed] [Google Scholar]
- Eisenbach M. Functions of the flagellar modes of rotation in bacterial motility and chemotaxis. Mol Microbiol. 1990 Feb;4(2):161–167. doi: 10.1111/j.1365-2958.1990.tb00584.x. [DOI] [PubMed] [Google Scholar]
- Goldman D. J., Ordal G. W. In vitro methylation and demethylation of methyl-accepting chemotaxis proteins in Bacillus subtilis. Biochemistry. 1984 Jun 5;23(12):2600–2606. doi: 10.1021/bi00307a010. [DOI] [PubMed] [Google Scholar]
- Helmann J. D., Chamberlin M. J. DNA sequence analysis suggests that expression of flagellar and chemotaxis genes in Escherichia coli and Salmonella typhimurium is controlled by an alternative sigma factor. Proc Natl Acad Sci U S A. 1987 Sep;84(18):6422–6424. doi: 10.1073/pnas.84.18.6422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S. Unidirectional digestion with exonuclease III creates targeted breakpoints for DNA sequencing. Gene. 1984 Jun;28(3):351–359. doi: 10.1016/0378-1119(84)90153-7. [DOI] [PubMed] [Google Scholar]
- Iino T., Komeda Y., Kutsukake K., Macnab R. M., Matsumura P., Parkinson J. S., Simon M. I., Yamaguchi S. New unified nomenclature for the flagellar genes of Escherichia coli and Salmonella typhimurium. Microbiol Rev. 1988 Dec;52(4):533–535. doi: 10.1128/mr.52.4.533-535.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones C. J., Macnab R. M. Flagellar assembly in Salmonella typhimurium: analysis with temperature-sensitive mutants. J Bacteriol. 1990 Mar;172(3):1327–1339. doi: 10.1128/jb.172.3.1327-1339.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kihara M., Homma M., Kutsukake K., Macnab R. M. Flagellar switch of Salmonella typhimurium: gene sequences and deduced protein sequences. J Bacteriol. 1989 Jun;171(6):3247–3257. doi: 10.1128/jb.171.6.3247-3257.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komeda Y., Kutsukake K., Iino T. Definition of additional flagellar genes in Escherichia coli K12. Genetics. 1980 Feb;94(2):277–290. doi: 10.1093/genetics/94.2.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komeda Y. Transcriptional control of flagellar genes in Escherichia coli K-12. J Bacteriol. 1986 Dec;168(3):1315–1318. doi: 10.1128/jb.168.3.1315-1318.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo S. C., Koshland D. E., Jr Sequence of the flaA (cheC) locus of Escherichia coli and discovery of a new gene. J Bacteriol. 1986 Jun;166(3):1007–1012. doi: 10.1128/jb.166.3.1007-1012.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutsukake K., Ohya Y., Iino T. Transcriptional analysis of the flagellar regulon of Salmonella typhimurium. J Bacteriol. 1990 Feb;172(2):741–747. doi: 10.1128/jb.172.2.741-747.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyte J., Doolittle R. F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982 May 5;157(1):105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Lipman D. J., Pearson W. R. Rapid and sensitive protein similarity searches. Science. 1985 Mar 22;227(4693):1435–1441. doi: 10.1126/science.2983426. [DOI] [PubMed] [Google Scholar]
- Magariyama Y., Yamaguchi S., Aizawa S. Genetic and behavioral analysis of flagellar switch mutants of Salmonella typhimurium. J Bacteriol. 1990 Aug;172(8):4359–4369. doi: 10.1128/jb.172.8.4359-4369.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malakooti J., Komeda Y., Matsumura P. DNA sequence analysis, gene product identification, and localization of flagellar motor components of Escherichia coli. J Bacteriol. 1989 May;171(5):2728–2734. doi: 10.1128/jb.171.5.2728-2734.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirel D. B., Chamberlin M. J. The Bacillus subtilis flagellin gene (hag) is transcribed by the sigma 28 form of RNA polymerase. J Bacteriol. 1989 Jun;171(6):3095–3101. doi: 10.1128/jb.171.6.3095-3101.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Márquez L. M., Helmann J. D., Ferrari E., Parker H. M., Ordal G. W., Chamberlin M. J. Studies of sigma D-dependent functions in Bacillus subtilis. J Bacteriol. 1990 Jun;172(6):3435–3443. doi: 10.1128/jb.172.6.3435-3443.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnishi K., Kutsukake K., Suzuki H., Iino T. Gene fliA encodes an alternative sigma factor specific for flagellar operons in Salmonella typhimurium. Mol Gen Genet. 1990 Apr;221(2):139–147. doi: 10.1007/BF00261713. [DOI] [PubMed] [Google Scholar]
- Ordal G. W., Nettleton D. O., Hoch J. A. Genetics of Bacillus subtilis chemotaxis: isolation and mapping of mutations and cloning of chemotaxis genes. J Bacteriol. 1983 Jun;154(3):1088–1097. doi: 10.1128/jb.154.3.1088-1097.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ordal G. W., Parker H. M., Kirby J. R. Complementation and characterization of chemotaxis mutants of Bacillus subtilis. J Bacteriol. 1985 Nov;164(2):802–810. doi: 10.1128/jb.164.2.802-810.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkinson J. S., Parker S. R., Talbert P. B., Houts S. E. Interactions between chemotaxis genes and flagellar genes in Escherichia coli. J Bacteriol. 1983 Jul;155(1):265–274. doi: 10.1128/jb.155.1.265-274.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T., Iino T., Horiguchi T., Yamaguchi S. Incomplete flagellar structures in nonflagellate mutants of Salmonella typhimurium. J Bacteriol. 1978 Feb;133(2):904–915. doi: 10.1128/jb.133.2.904-915.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabor S., Richardson C. C. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc Natl Acad Sci U S A. 1985 Feb;82(4):1074–1078. doi: 10.1073/pnas.82.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoelke M. S., Casper J. M., Ordal G. W. Methyl group turnover on methyl-accepting chemotaxis proteins during chemotaxis by Bacillus subtilis. J Biol Chem. 1990 Feb 5;265(4):1928–1932. [PubMed] [Google Scholar]
- Thoelke M. S., Kirby J. R., Ordal G. W. Novel methyl transfer during chemotaxis in Bacillus subtilis. Biochemistry. 1989 Jun 27;28(13):5585–5589. doi: 10.1021/bi00439a037. [DOI] [PubMed] [Google Scholar]
- Thoelke M. S., Parker H. M., Ordal E. A., Ordal G. W. Rapid attractant-induced changes in methylation of methyl-accepting chemotaxis proteins in Bacillus subtilis. Biochemistry. 1988 Nov 1;27(22):8453–8457. doi: 10.1021/bi00422a024. [DOI] [PubMed] [Google Scholar]
- Yamaguchi S., Aizawa S., Kihara M., Isomura M., Jones C. J., Macnab R. M. Genetic evidence for a switching and energy-transducing complex in the flagellar motor of Salmonella typhimurium. J Bacteriol. 1986 Dec;168(3):1172–1179. doi: 10.1128/jb.168.3.1172-1179.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuberi A. R., Ying C. W., Parker H. M., Ordal G. W. Transposon Tn917lacZ mutagenesis of Bacillus subtilis: identification of two new loci required for motility and chemotaxis. J Bacteriol. 1990 Dec;172(12):6841–6848. doi: 10.1128/jb.172.12.6841-6848.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuberi A. R., Ying C. W., Weinreich M. R., Ordal G. W. Transcriptional organization of a cloned chemotaxis locus of Bacillus subtilis. J Bacteriol. 1990 Apr;172(4):1870–1876. doi: 10.1128/jb.172.4.1870-1876.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]