Abstract
The DNA-dependent protein kinase (DNA-PK) is required for DNA double-strand break (DSB) repair and immunoglobulin gene rearrangement and may play a role in the regulation of transcription. The DNA-PK holoenzyme is composed of three polypeptide subunits: the DNA binding Ku70/86 heterodimer and an ≈460-kDa catalytic subunit (DNA-PKcs). DNA-PK has been hypothesized to assemble at DNA DSBs and play structural as well as signal transduction roles in DSB repair. Recent advances in atomic force microscopy (AFM) have resulted in a technology capable of producing high resolution images of native protein and protein–nucleic acid complexes without staining or metal coating. The AFM provides a rapid and direct means of probing the protein–nucleic acid interactions responsible for DNA repair and genetic regulation. Here we have employed AFM as well as electron microscopy to visualize Ku and DNA-PK in association with DNA. A significant number of DNA molecules formed loops in the presence of Ku. DNA looping appeared to be sequence-independent and unaffected by the presence of DNA-PKcs. Gel filtration of Ku in the absence and the presence of DNA indicates that Ku does not form nonspecific aggregates. We conclude that, when bound to DNA, Ku is capable of self-association. These findings suggest that Ku binding at DNA DSBs will result in Ku self-association and a physical tethering of the broken DNA strands.
Keywords: DNA repair, atomic force microscopy, electron microscopy, double-strand break
The repair of DNA double-strand breaks (DSBs) is required for cellular survival after exposure to ionizing radiation as well as the normal rearrangement of immunoglobulin and T cell receptor genes in lymphocytes (1–3). Therefore, DSB repair is critical to normal development of the immune system and the maintenance of genomic integrity. Left unrepaired DSBs can prove lethal or mutagenic. Differences in DSB repair between yeast and higher eukaryotic species were initially uncovered during early gene targeting experiments in mammalian cells. In yeast, DSB repair occurs largely by homologous recombination (4–7). Although capable of homologous recombination, higher eukaryotic species repair DSBs primarily by a process of nonhomologous or illegitimate recombination (8–13).
Illegitimate recombination joins DNA strands that share little or no sequence homology via an end-to-end joining mechanism (14–17). This process is capable of preserving the sequences of a variety of end structures (15, 17). One model proposed to explain these findings includes an end alignment protein that acts at the break site to allow fill-in synthesis by a polymerase at problematic break sites such as 3′ overhangs (17). This model may also address the problem of holding or tethering broken ends in place to help ensure their physical proximity for correct repair. However, simple tethering and the presumably more demanding alignment of broken ends that would allow fill-in synthesis need not be provided by the same protein. An early step in DSB repair may be a tethering of broken DNA strands while other repair factors are recruited to the break site. In this case, the actual “alignment” of ends postulated by Thode et al. (17) could be provided by other members of the repair apparatus or by the organization of DNA in nucleosomes and chromatin. In theory, the tethering of broken DNA strands could also be accomplished by the self-association of DNA end-binding proteins.
Although little is known about the mechanism of DSB repair, genetic studies have revealed that the DNA-dependent protein kinase (DNA-PK) is required for the process in mammalian cells (18–28). The biological role of DNA-PK in DSB repair remains to be determined, but a number of characteristics have led to the hypothesis that the serine/threonine kinase plays a structural as well as a signal transduction role. Proposed structural roles include tethering DNA ends and providing a scaffolding for the assembly of a repair complex (2, 3, 29).
The DNA-PK holoenzyme is formed by association of the ≈460-kDa catalytic subunit (DNA-PKcs) with a DNA binding component known as Ku (30–34). Ku is a heterodimer of 70-kDa and 86-kDa subunits that is capable of both sequence-independent and sequence-specific DNA binding. While sequence-specific binding may be involved in transcriptional regulation (35–43), sequence-independent binding is consistent with a role in DSB repair. Ku requires free DNA ends, hairpin loops, single-strand nicks, or gaps for sequence-independent DNA binding (44–46). The requirement for Ku and DNA-PK in the DSB repair pathway taken together with the affinity of Ku for free DNA ends suggest these proteins as candidate end-tethering factors (2, 3, 29).
A role for Ku or DNA-PK in DNA end-tethering predicts either self-association of the DNA-bound proteins or a capacity for the complex to bind two DNA ends simultaneously. To directly examine the behavior of Ku and DNA-PK in association with linear DNA we have used a combination of electron and atomic force microscopy (AFM). AFM allows the visualization of unstained and uncoated protein–nucleic acid complexes at high resolution. With the ability to image specimens in air, aqueous buffers, or organic solvents, AFM is uniquely suited for biological studies and provides a means by which the macromolecular interactions responsible for genetic and cellular regulation can be characterized under native conditions. The studies of Ku and DNA-PK presented here indicate that DNA-bound Ku mediates the formation of DNA loops. Addition of DNA-PKcs to Ku-bound DNA forms a large DNA-bound complex both at DNA ends and internal positions. The looping behavior observed for Ku is unaffected by the presence of DNA-PKcs.
MATERIALS AND METHODS
Purification of Ku and DNA-PKcs from HeLa cells.
Ku70/86 was purified from HeLa cell nuclear extracts using an anti-Ku80 antibody affinity column. Antibodies specific for Ku86 (47) were conjugated to protein G agarose beads using dimethylpimelimidate HCl to achieve a final concentration of 1 mg antibody/ml. HeLa cell nuclear extract (50 ml) (48) containing 500 mg of protein was incubated with 2 ml of the anti-Ku80 matrix for 4 hr at 4°C in the presence of 100 μg/ml sheared salmon sperm DNA. The resin was then packed into a 5-ml disposal column (Bio-Rad) and washed sequentially with 5 column volumes of Hepes chromatography buffer (50 mM Hepes, pH 7.5/2 mM EDTA/0.01% Nonidet P-40/1 mM DTT/20 μg/ml phenylmethylsulfonyl fluoride/1 μg/ml aprotinin/pepstatin A/leupeptin) containing 0.1 M, 0.2 M, and 0.5 M KCl. Ku70/86 was eluted from the washed column matrix using 5 column volumes of 50 mM Tris⋅HCl, pH 7.9/50% ethylene glycol/1.75 M MgCl2. Ku70/86 was further purified by heparin agarose chromatography. Ku70/86 was loaded on a 1-ml Hi-Trap heparin agarose column (Pharmacia) in Hepes chromatography buffer containing 0.1 M KCl. Proteins were eluted from the column sequentially by addition of Hepes chromatography buffer containing 0.1, 0.2, 0.3, 0.5, and 1.0 M KCl. Ku70/86 eluted from the heparin agarose column in the 0.3 M KCl fraction. The fraction was dialyzed in TM buffer containing 0.1 M KCl and frozen in aliquots at −70°C. The protein concentration of the affinity purified Ku70/86 was determined by Bradford analysis using bovine serum albumin as a standard. Recombinant Ku70/86 was purified from Sf9 cell extracts using SP-Sepharose, Q-Sepharose, heparin agarose, and DNA-affinity chromatography as described (J.W., unpublished data).
DNA-PKcs was purified from HeLa cell nuclear extract by sequential ion-exchange chromatography using phosphocellulose P-11, DEAE-Sepharose, and heparin agarose columns and a Superdex-200 gel filtration column as described (48). Superdex-200 fractions containing DNA-PKcs were then incubated for 1 hr at 4°C with 1 ml anti-Ku80 affinity resin that was preabsorbed with 1 mg of Ku70/86 in Hepes chromatography buffer containing 0.1 M KCl and 100 μg/ml sheared salmon sperm DNA. DNA-PKcs was eluted from the Ku80 affinity matrix in 5 ml of Hepes chromatography buffer containing 0.5 M KCl. The ionic strength of the eluted DNA-PKcs solution was then adjusted to 0.1 M KCl and applied to a 1-ml Hi-Trap Q-Sepharose column (Pharmacia). DNA-PKcs was eluted from the column in Hepes chromatography buffer containing 0.3 M KCl. Prior to use the DNA-PKcs was dialyzed in TM buffer containing 0.1 M KCl. The concentration of the affinity-purified DNA-PKcs was determined by Bradford analysis using bovine serum albumin as a standard.
Preparation of DNA Substrates.
pUC19 was purchased from New England Biolabs. Digestion with SmaI to generate a linearized 2.6-kb DNA fragment was performed under standard conditions. Digested DNA was purified by phenol/chloroform extraction followed by gel band isolation using the QIAEX II gel purification kit (Qiagen, Chatsworth, CA). The 660-bp DNA fragment used for statistical analysis was generated by PCR using primers complimentary to the region of the Ku86 cDNA encoding amino acids 1–220 (49). This coding sequence was selected for two reasons: (i) the length of the fragment was such that spontaneous loop formation would be low, and (ii) the sequence does not contain any known protein binding motifs. The PCR product was purified by phenol/chloroform extraction and gel band isolation.
Electron and Atomic Force Microscopy.
All reactions were performed in 50 mM Hepes, pH 7.5/100 mM KCl/10 mM MgCl2/1 mM DTT/0.1 mM EDTA (Hepes buffer). DNA–Ku complexes for electron microscopy were formed by incubation of 5 μg/ml SmaI-linearized pUC19 and 5 μg/ml Ku in a volume of 100 μl for 20 min at 37°C. DNA–PK complexes were formed by the further addition of 12 μg/ml DNA-PKcs. After chemical fixation with 0.6% glutaraldehyde, reaction mixtures were chromatographed on a 2-ml bed volume of Bio-Gel A-15m (Bio-Rad). Column fractions were mounted by the spermidine mounting technique, dehydrated in a graded series of ethanol solutions, and rotary shadow cast with tungsten (50, 51). Samples were imaged and photographed on a Philips CM30 at an accelerating voltage of 50 kV.
For atomic force microscopy, samples were prepared from 10-μl reactions containing 0.4 μg/ml pUC19 DNA or 0.1 μg/ml 660-bp DNA in Hepes buffer equivalent to approximately 0.25 nM DNA. Ku was used at a final concentration of 1 μg/ml (≈6.5 nM), whereas DNA-PKcs was used at final concentration of 2 μg/ml (≈4 nM). Reactions were mounted by adsorption to freshly cleaved mica in the presence of 10 mM MgCl2. Mounted specimens were fixed using 0.6% glutaraldehyde unless otherwise indicated. Prior to imaging, samples were dehydrated in a graded series of ethanol solutions. Atomic force microscopy was performed using tapping mode in air on a NanoScope III (Digital Instruments, Santa Barbara, CA).
Statistical Analysis.
For statistical analysis all samples contained 0.1 μg/ml 660-bp linear DNA. When present Ku was used at a concentration of 1 μg/ml, DNA-PKcs at 2 μg/ml, and ATP at a final concentration of 6 mM. After incubation for 30 min at 30°C, reactions were mounted unfixed on freshly cleaved mica. Molecules were imaged and scored as linear, looped, or multi-DNA complexes using 2-μm AFM scans of random areas. Frequency and χ2 statistics were performed using spss/mac (SPSS, Chicago) on a Power Macintosh 9500/132 (Apple, Cupertino, CA).
Gel Filtration Chromatography.
For gel filtration analysis of recombinant Ku, 600 pmol of Ku70/86 in a final volume of 750 μl was applied to a HiLoad Superdex 200 16/60 (Pharmacia) gel filtration column at a flow rate of 1 ml/min. One-milliliter fractions were collected at the same flow rate and the absorbance of eluted fractions measured at 280 nm. Gel filtration analysis of Ku/DNA complexes was performed identically with the exception that 600 pmol of recombinant Ku70/86 was incubated with 600 pmol of a 24-bp double-stranded DNA fragment of the sequence 5′-GAGCTCGGTACCCGGGATCCTCTA-3′ in a final volume of 750 μl at 25°C for 20 min prior to loading on the Superdex 200 column. High molecular weight gel filtration standards (Pharmacia) were chromatographed under identical conditions to calibrate the column. For silver-stain analysis of the column profile, 10 μl of each sample was resolved by 10% SDS/PAGE and silver-stained as described previously (52).
Electrophoretic Mobility Shift Assay (EMSA).
The same DNA fragment used in the gel filtration experiment was radiolabeled using 32P[γ-ATP] and T4 polynucleotide kinase. For EMSAs 10 pmol of the labeled DNA was incubated with various amounts of Ku, as indicated in the appropriate figure legend, for 20 min at 25°C in a final volume of 20 μl. Reaction products were analyzed by PAGE using 0.5× TAE (20 mM Tris acetate/500 μM EDTA) running buffer.
RESULTS
Visualization of Ku and DNA-PK Bound to Linear DNA.
Human Ku heterodimer was purified from HeLa cells and baculovirus-transformed insect cells (Fig. 1). The recombinant Ku was purified to >95% purity. Ku–DNA complexes for atomic force microscopy were formed by incubating 1 μg/ml HeLa-purified or recombinant Ku with 0.4 μg/ml SmaI-linearized pUC19 DNA. Time course experiments performed by incubation on ice revealed that recombinant and HeLa-purified Ku behaved similarly to each other and to previous microscopic analysis of Ku in the presence of linear DNA (ref. 53; data not shown).
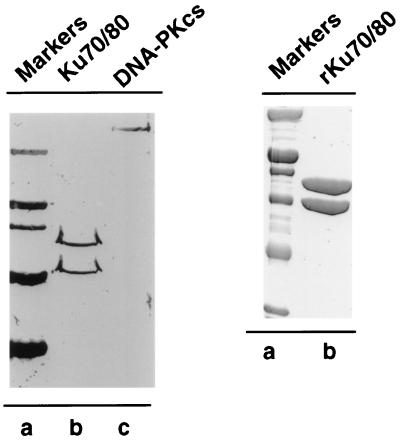
Figure 1.
SDS/PAGE analysis of HeLa DNA-PK and recombinant Ku70/86. (A) 100 ng of Ku70/86 (lane b) and 100 ng of DNA-PKcs (lane c) purified from HeLa cell nuclear extract was resolved by 10% SDS/PAGE and analyzed by silver staining. Molecular mass markers (lane a) are as follows: Myosin (200 kDa), β-galactosidase (116 kDa), phosphorylase B (97.4 kDa), serum albumin (66.2 kDa), and ovalbumin (45.0 kDa). (B) Purified recombinant Ku70/86 (5 μg) (lane b) was resolved by 10% SDS/PAGE and analyzed by Coomassie blue staining. Molecular mass markers (lane a) are as follows: Myosin (200 kDa), β-galactosidase (116 kDa), phosphorylase B (97.4 kDa), serum albumin (66.2 kDa), ovalbumin (45.0 kDa), and carbonic anhydrase (31.0 kDa).
DNA-PKcs was purified from HeLa cells and examined by atomic force and electron microscopy. In the absence of added Ku, association between DNA and DNA-PKcs was relatively rare (19%, n = 177; Fig. 2A). Binding of DNA-PKcs to DNA in the absence of Ku may reflect a weak interaction of the catalytic subunit with DNA; however, the presence of small amounts of contaminating Ku may also be a factor. The addition of recombinant Ku to reactions containing DNA and DNA-PKcs resulted in a marked increase in the number of DNA fragments associated with the large DNA-PK subunit (77%, n = 71; Fig. 2B). Although not all DNA-bound Ku were associated with DNA-PKcs, the DNA-PK was not localized exclusively to the DNA ends. Internally bound as well as end-bound DNA-PK could easily be identified (Fig. 2 C–E). Free DNA-PKcs and Ku particles present on the mica surface served as internal size standards, allowing Ku and DNA-PK to be distinguished. The identification of DNA-PK at internal positions indicates that either the DNA–PK complex is capable of translocation or that the DNA-PKcs is capable of association with internally bound Ku. These alternatives are not mutually exclusive, and a combination of both behaviors may be responsible for the observed distribution of DNA-PK.
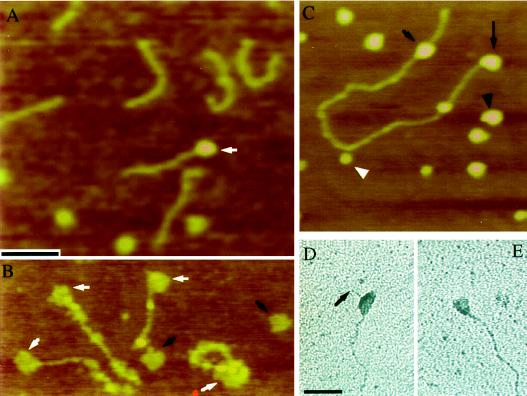
Figure 2.
AFM and electron microscopy of DNA-PK complexes. (A) Reactions containing DNA-PKcs and a linear 660-bp DNA fragment were examined by AFM. Association between DNA-PKcs and DNA (arrow) was observed but uncommon compared with reactions that also contained Ku. (B) In the presence of recombinant Ku, DNA-PKcs was frequently seen associated with the DNA fragment (white arrows). Black arrows indicate unbound DNA-PKcs. Samples in A and B were mounted and imaged without fixation. (C) Linearized pUC19 incubated in the presence of Ku and DNA-PKcs. Size differences allowed the DNA-PKcs (black arrowhead) to be distinguished from recombinant Ku particles (white arrowhead). End-bound (long arrow) and internally bound DNA-PK (short arrow) were both commonly observed. (D and E) Electron microscopy of rotary-shadowed DNA-PK/DNA complexes assembled using HeLa-purified Ku revealed similar DNA-PK behavior. (D) A DNA-PK complex bound internally to linearized pUC19. The arrow indicates the DNA end. (E) An example of an end-bound DNA-PK complex. Bar in A is 100 nm for A–C. Bar in D is 100 nm for D and E.
Loops Are Associated with Ku DNA Binding.
Ku particles exhibited a tendency to cluster along the DNA (e.g., Fig. 3). It was not clear whether this effect was due to translocation effects or self-association. Qualitative examination of a large number of molecules from reactions containing Ku and DNA suggested the protein may promote DNA loop formation (Fig. 3 A–D). To determine whether there was a statistically significant looping of DNA mediated by Ku or DNA-PK, we scored micrographs of a 660-bp DNA fragment, incubated in the absence or presence of Ku (Fig. 3 E–I) or Ku and DNA-PKcs (Fig. 3 J–M). We used a short DNA fragment instead of pUC19 DNA to reduce the number of spontaneous loops formed by folding during the mounting procedure. Recombinant Ku was judged to be of higher purity than material purified from HeLa cells and was therefore used for all statistical analyses.
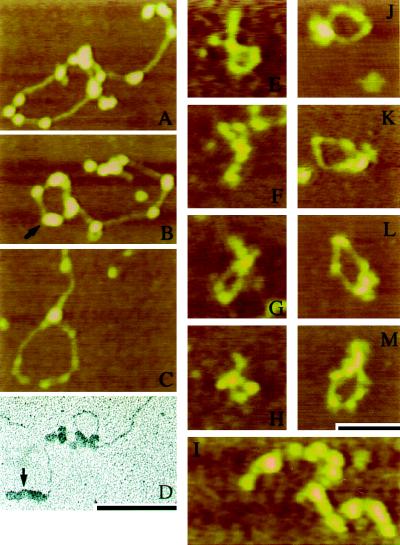
Figure 3.
Qualitative examination of linearized pUC19 incubated with either recombinant Ku (A and B) or HeLa Ku (C and D) revealed the formation of Ku-associated loops. Clusters of Ku particles were common (arrows in B and D). For statistical analysis, a 660-bp DNA fragment was used to quantitate the looping effect initially observed for Ku and DNA-PK on linearized pUC19. For these studies, all samples were mounted and imaged without fixation. (E–H) Examples of looped 660-bp DNA formed in the presence of recombinant Ku. (I) Multi-DNA complexes were also observed at an increased frequency in the presence of recombinant Ku. (J–M) Loops formed in the presence of Ku and the DNA-PKcs. A free DNA-PKcs particle is visible in the lower right corner of J. Bar in M is 100 nm for A–C and E–M. Bar in D is 100 nm for D.
Spontaneous looping was rare (2%) in reactions containing the 660-bp DNA alone (Table 1). At the DNA concentration used here, the density of DNA on the mica surface was such that few DNA molecules were seen touching. The addition of 1 μg/ml Ku resulted in 17% looped DNA fragments and 5.5% multi-DNA complexes; a χ2 test indicates these results are significantly different from DNA-only controls (P < 0.001).
Table 1.
AFM analysis of molecules from reactions containing a 660-bp DNA fragment alone or in the presence of DNA-PK components
| DNA | +Ku | +DNA-PK | DNA-PK + ATP | |
|---|---|---|---|---|
| Looped | 2.3% | 17.3% | 16.5% | 11.2% |
| (7) | (38) | (43) | (31) | |
| Multi-DNA complex | 1.3% | 5.5% | 4.6% | 4.7% |
| (4) | (12) | (12) | (13) | |
| Linear | 96.4% | 77.3% | 78.8% | 84.1% |
| (292) | (170) | (205) | (233) | |
| n | 303 | 220 | 260 | 277 |
For statistical analysis, reactions were mounted unfixed and examined by AFM. The images were scored for the presence of linear, looped, or multiple DNA complexes. Reactions contained a 660-bp linear DNA fragment either alone (DNA), or in the presence of recombinant Ku (+Ku), recombinant Ku and DNA-PKcs (+DNA-PK), or recombinant Ku and DNA-PKcs plus ATP (DNA-PK + ATP). The percentage of reaction totals are followed by the number of molecules observed in parentheses.
To examine the effect of DNA-PKcs on DNA looping mediated by Ku, micrographs of the 660-bp linear DNA in the presence of Ku and the DNA-PKcs were scored for the presence of looped or multi-DNA structures. Loops in the presence of DNA-PKcs were formed with a similar frequency to those formed in the presence of Ku, indicating the presence of the DNA-PK complex neither inhibited nor stimulated the formation of DNA loops.
DNA-PK autophosphorylation leads to inactivation of DNA-PK and dissociation of DNA-PKcs from Ku (54). Ku is also known to be a substrate of DNA-PK in vitro (55). It was therefore of interest to determine whether the presence of ATP and DNA-PKcs effected the formation of DNA loops. The same DNA, Ku, and DNA-PKcs concentrations as for the Ku and Ku + DNA-PKcs experiments described above were used for reactions carried out in the presence of 6 mM ATP. These reactions were performed and mounted in parallel with the control, Ku, and DNA-PK reactions described above. A decrease in the number of DNA-PK/DNA complexes was evident in reactions containing ATP, consistent with DNA-PK autophosphorylation resulting in dissociation of DNA-PKcs from Ku; nonetheless, DNA-PK-associated DNA was still observed. Similar to reactions containing Ku or DNA-PK, the ATP-containing reaction was significantly different from DNA alone (P < 0.001). Although the DNA-PK + ATP reaction was not significantly different from Ku or DNA-PK reactions by a χ2 test (P < 0.13), loop formation was less frequent (11%).
Gel Filtration of Free and DNA-Bound Ku.
Ku-mediated DNA loops could result from a nonspecific aggregation of Ku. This possibility seemed unlikely given that free Ku particles observed on mica under the AFM were of a uniform size and did not demonstrate a tendency to aggregate. Nonetheless, it is possible that DNA-bound Ku may aggregate. To address this issue, Superdex-200 gel filtration chromatography was used to examine the behavior of Ku in the presence and absence of the 24-bp oligonucleotide. Under the conditions used for gel filtration, the oligonucleotide was shown by an EMSA to bind only a single Ku (Fig. 4A). Peak Ku70/86 eluted from the column in the absence of DNA between fractions 65 and 69 (Fig. 4B). In the presence of the 24-bp DNA fragment, peak Ku70/86 eluted in fractions 59–63. The apparent molecular mass of Ku in the absence of DNA was determined to be 158 kDa. However, in the presence of the 24-bp DNA fragment Ku eluted as a 325-kDa species based on the elution characteristics relative to aldolase (158 kDa), catalase (232 kDa), ferritin (440 kDa), and thyroglobulin (669 kDa). Larger species, which would be predicted if DNA-bound Ku formed nonspecific aggregates, were not observed. The elution profile of the oligonucleotide alone was consistent with a small molecule outside the resolving range of the Superdex-200 column (data not shown).
Figure 4.
DNA-binding and gel filtration analysis of recombinant Ku70/86. (A) Electrophoretic mobility shift analysis of recombinant Ku70/86. DNA (10 pmol) was incubated with the indicated amounts of Ku70/86 and analyzed by gel electrophoresis as described. Results were obtained by autoradiography of the dried gel. (B) Samples from fractions 59–72 of Superdex 200 column chromatography runs of Ku70/86 alone or Ku70/86 in the presence of a 24-bp double-stranded DNA fragment were resolved by 10% SDS/PAGE and analyzed by silver staining. Molecular mass markers (lane M) are as follows: Myosin (200 kDa), β-galactosidase (116 kDa), phosphorylase B (97.4 kDa), serum albumin (66.2 kDa), ovalbumin (45.0 kDa), and carbonic anhydrase (31.0 kDa).
DISCUSSION
The AFM provides a powerful tool for addressing aspects of protein–nucleic acid interactions difficult to study by traditional biochemical methods. In particular, application of AFM to problems in DNA repair and gene regulation promises to unlock aspects of these processes that are not easily addressed by more conventional approaches. Here we have applied AFM to determine whether a statistically significant fraction of Ku- or DNA-PK-bound DNAs display loop formation, a state indicative of protein–protein interaction by the DNA-bound proteins. DNA loops were formed in the presence of Ku at a frequency of ≈10–15%, a relatively high percentage for in vitro DNA looping (e.g., compare with refs. 56–58). No obvious sequence specificity for the looping effect was detected; loops formed in an apparently random distribution along the length of pUC19 DNA as well as a short, unrelated sequence. Addition of DNA-PKcs to Ku–DNA binding reactions did not affect the frequency of loop formation. Based on the size of particles observed at loop junctions, Ku-mediated loops resulted from the association of two or more heterodimers. DNA-bound Ku not participating in loop formation was often found as a group of two or more Ku particles (Fig. 3), suggesting that self-association takes place in the presence of DNA. However, we found no evidence that Ku forms nonspecific aggregates.
Gel filtration of Ku in the absence of DNA indicates the Ku particle fractionates in a manner consistent with a 158-kDa particle (Fig. 4B; ref. 59). In the presence of an oligonucleotide, shown to bind only a single Ku by EMSA (Fig. 4A), the elution profile changes dramatically. Under these conditions, the majority of Ku elutes in a fraction corresponding to a molecular mass of approximately 325 kDa. Considering the elution profile of the oligonucleotide, this increase in size does not appear to result simply from association of Ku with the 24-bp DNA. The effect could be explained by a conformational change in Ku upon binding to DNA; however, the fact that the size corresponds closely with the predicted molecular mass of two Ku heterodimers suggests that the species may represent Ku tetramers. The formation of Ku tetramers in the presence of DNA was previously suggested to explain the behavior of Ku when bound to the CTC box of the collagen IV promoter (60). The formation of tetrameric species in the presence of DNA is consistent with the looping behavior observed by microscopic analysis. Gel filtration was performed at Ku and DNA concentrations approximately two orders of magnitude higher than those used for microscopy. If the looping phenomenon was a result of nonspecific aggregation we would expect aggregates to be detected during gel filtration.
Previous reports have found Ku capable of binding to linear DNA ends and translocating to internal positions with little or no sequence specificity (53). Ku is capable of binding to hairpin structures, single-stranded nicks, and gaps in an apparently sequence-independent manner (44–46, 61). A number of studies have also suggested that the Ku protein is capable of sequence-specific DNA binding (36–40, 60). Therefore, there appear to be two distinct modes of Ku–DNA interaction: a sequence-independent and a sequence-dependent mode. Transcriptional regulation by DNA-PK is consistent with sequence-specific interaction with regulatory elements. DNA damage recognition, however, is more likely to require the sequence-independent end-loading behavior of Ku.
Sequence-independent DNA looping by Ku is consistent with a dual role for Ku in transcription and DSB repair. DNA looping is a well established phenomenon that may play a role in the regulation of transcription, DNA replication, and recombination (62, 63). A common example is loop formation by the self-association of transcription factors bound at two distant sites (56–58). The looping mediated by Ku, however, does not appear to be localized to specific binding sites because the two unrelated DNA sequences examined here both display Ku-associated loops. In vivo it is likely that, in the absence of DSBs or nicks, Ku must associate with chromatin via sequence-specific binding. In this case, Ku-mediated looping would take on sequence-specific characteristics and may place activated DNA-PK at sites where DNA bound substrates can be phosphorylated. In vivo, chromosomes provide a far more complex substrate for Ku and DNA-PK association than the naked DNA fragments used in our microscopic study. The ability of Ku to translocate from DNA ends to internal positions could be inhibited in vivo, either partially or completely, by the presence of other DNA-associated complexes. Thus, in vivo, DSB-bound Ku may be restricted to DNA ends. If so, the self-association resulting in loop formation in vitro could result in end-tethering in vivo. Under the conditions employed in this study we observed intermolecular association mediated by Ku at a much lower frequency than intramolecular interactions, consistent with the relatively low DNA concentrations used favoring intramolecular associations (62). The high local concentration of DNA ends present at DSBs in situ would presumably favor intermolecular Ku association. If this is the case, Ku may provide an initial tethering of DNA ends after the formation of DSBs. Subsequently, other protein factors may associate with the Ku/DNA-PK complex to provide further end alignment and repair functions.
Acknowledgments
We thank Mateo Aragón for technical assistance, Lonna Atkeson for assistance with statistical analyses, Marilyn Hawley of the Center for Materials Science (Los Alamos National Laboratory, Los Alamos, NM) for providing access to an AFM during pilot studies, and Harriet Kung of Materials Science and Technology division for providing access to the Materials Science Laboratory electron microscope facility at Los Alamos. This work was supported by the U.S. Department of Energy and by National Institutes of Health Grant CA50519 to D.J.C. and by National Science Foundation Grant MCB-9408231 to D.G.B. Use of the University of New Mexico electron microscopes was paid for by the University of New Mexico Dedicated Health Research Fund. R.B.C. is supported by an Alexander Hollaender Distinguished Postdoctoral Fellowship sponsored by the U.S. Department of Energy, Office of Health and Environmental Research, and administered by the Oak Ridge Institute for Science and Education.
ABBREVIATIONS
- DSB
double-strand break
- DNA-PK
DNA-dependent protein kinase
- DNA-PKcs
DNA-PK catalytic subunit
- AFM
atomic force microscopy
- EMSA
electrophoretic mobility shift assay
References
- 1.Alt F W, Oltz E M, Young F, Gorman J, Taccioli G, Chen J. Immunol Today. 1992;13:306–314. doi: 10.1016/0167-5699(92)90043-7. [DOI] [PubMed] [Google Scholar]
- 2.Jackson S P, Jeggo P A. Trends Biochem Sci. 1995;20:412–415. doi: 10.1016/s0968-0004(00)89090-8. [DOI] [PubMed] [Google Scholar]
- 3.Jeggo P A, Taccioli G E, Jackson S P. Bioessays. 1995;17:949–957. doi: 10.1002/bies.950171108. [DOI] [PubMed] [Google Scholar]
- 4.Hinnen A, Hicks J B, Fink G R. Proc Natl Acad Sci USA. 1978;75:1929–1933. doi: 10.1073/pnas.75.4.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Orr-Weaver T L, Szostak J W, Rothstein R J. Proc Natl Acad Sci USA. 1981;78:6354–6358. doi: 10.1073/pnas.78.10.6354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Orr-Weaver T L, Szostak J W. Proc Natl Acad Sci USA. 1983;80:4417–4421. doi: 10.1073/pnas.80.14.4417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rothstein R. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- 8.Roth D B, Wilson J H. Proc Natl Acad Sci USA. 1985;82:3355–3359. doi: 10.1073/pnas.82.10.3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Robins D M, Ripley S, Henderson A S, Axel R. Cell. 1981;23:29–39. doi: 10.1016/0092-8674(81)90267-1. [DOI] [PubMed] [Google Scholar]
- 10.Lin F L, Sperle K, Sternberg N. Proc Natl Acad Sci USA. 1985;82:1391–1395. doi: 10.1073/pnas.82.5.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smith A J, Berg P. Cold Spring Harbor Symp Quant Biol. 1984;49:171–181. doi: 10.1101/sqb.1984.049.01.020. [DOI] [PubMed] [Google Scholar]
- 12.Smithies O, Gregg R G, Boggs S S, Koralewski M A, Kucherlapati R S. Nature (London) 1985;317:230–234. doi: 10.1038/317230a0. [DOI] [PubMed] [Google Scholar]
- 13.Thomas K R, Folger K R, Capecchi M R. Cell. 1986;44:419–428. doi: 10.1016/0092-8674(86)90463-0. [DOI] [PubMed] [Google Scholar]
- 14.Pfeiffer P, Vielmetter W. Nucleic Acids Res. 1988;16:907–924. doi: 10.1093/nar/16.3.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roth D B, Wilson J H. Mol Cell Biol. 1986;6:4295–4304. doi: 10.1128/mcb.6.12.4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roth D B, Porter T N, Wilson J H. Mol Cell Biol. 1985;5:2599–2607. doi: 10.1128/mcb.5.10.2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thode S, Schafer A, Pfeiffer P, Vielmetter W. Cell. 1990;60:921–928. doi: 10.1016/0092-8674(90)90340-k. [DOI] [PubMed] [Google Scholar]
- 18.Boubnov N V, Hall K T, Wills Z, Lee S E, He D M, Benjamin D M, Pulaski C R, Band H, Reeves W, Hendrickson E A, Weaver D T. Proc Natl Acad Sci USA. 1995;92:890–894. doi: 10.1073/pnas.92.3.890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smider V, Rathmell W K, Lieber M R, Chu G. Science. 1994;266:288–291. doi: 10.1126/science.7939667. [DOI] [PubMed] [Google Scholar]
- 20.Blunt T, Finnie N J, Taccioli G E, Smith G C, Demengeot J, Gottlieb T M, Mizuta R, Varghese A J, Alt F W, Jeggo P A, Jackson S P. Cell. 1995;80:813–823. doi: 10.1016/0092-8674(95)90360-7. [DOI] [PubMed] [Google Scholar]
- 21.Finnie N J, Gottlieb T M, Blunt T, Jeggo P A, Jackson S P. Proc Natl Acad Sci USA. 1995;92:320–324. doi: 10.1073/pnas.92.1.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kirchgessner C U, Patil C K, Evans J W, Cuomo C A, Fried L M, Carter T, Oettinger M A, Brown J M. Science. 1995;267:1178–1183. doi: 10.1126/science.7855601. [DOI] [PubMed] [Google Scholar]
- 23.Peterson S R, Kurimasa A, Oshimura M, Dynan W S, Bradbury E M, Chen D J. Proc Natl Acad Sci USA. 1995;92:3171–3174. doi: 10.1073/pnas.92.8.3171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Taccioli G E, Gottlieb T M, Blunt T, Priestley A, Demengeot J, Mizuta R, Lehmann A R, Alt F W, Jackson S P, Jeggo P A. Science. 1994;265:1442–1445. doi: 10.1126/science.8073286. [DOI] [PubMed] [Google Scholar]
- 25.Lees-Miller S P, Godbout R, Chan D W, Weinfeld M, Day R S, III, Barron G M, Allalunis-Turner J. Science. 1995;267:1183–1185. doi: 10.1126/science.7855602. [DOI] [PubMed] [Google Scholar]
- 26.Rathmell W K, Chu G. Proc Natl Acad Sci USA. 1994;91:7623–7627. doi: 10.1073/pnas.91.16.7623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen F, Peterson S R, Story M D, Chen D J. Mutat Res. 1996;362:9–19. doi: 10.1016/0921-8777(95)00026-7. [DOI] [PubMed] [Google Scholar]
- 28.Finnie N J, Gottlieb T M, Blunt T, Jeggo P A, Jackson S P. Philos Trans R Soc London B. 1996;351:173–179. doi: 10.1098/rstb.1996.0014. [DOI] [PubMed] [Google Scholar]
- 29.Roth D B, Lindahl T, Gellert M. Curr Biol. 1995;5:496–499. doi: 10.1016/s0960-9822(95)00101-1. [DOI] [PubMed] [Google Scholar]
- 30.Lees-Miller S P, Chen Y R, Anderson C W. Mol Cell Biol. 1990;10:6472–6481. doi: 10.1128/mcb.10.12.6472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carter T, Vancurova I, Sun I, Lou W, DeLeon S. Mol Cell Biol. 1990;10:6460–6471. doi: 10.1128/mcb.10.12.6460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dvir A, Peterson S R, Knuth M W, Lu H, Dynan W S. Proc Natl Acad Sci USA. 1992;89:11920–11924. doi: 10.1073/pnas.89.24.11920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gottlieb T M, Jackson S P. Cell. 1993;72:131–142. doi: 10.1016/0092-8674(93)90057-w. [DOI] [PubMed] [Google Scholar]
- 34.Hartley K O, Gell D, Smith G C, Zhang H, Divecha N, Connelly M A, Admon A, Lees-Miller S P, Anderson C W, Jackson S P. Cell. 1995;82:849–856. doi: 10.1016/0092-8674(95)90482-4. [DOI] [PubMed] [Google Scholar]
- 35.Knuth M W, Gunderson S I, Thompson N E, Strasheim L A, Burgess R R. J Biol Chem. 1990;265:17911–17920. [PubMed] [Google Scholar]
- 36.Giffin W, Torrance H, Rodda D J, Prefontaine G G, Pope L, Hache R J. Nature (London) 1996;380:265–268. doi: 10.1038/380265a0. [DOI] [PubMed] [Google Scholar]
- 37.Roberts M R, Han Y, Fienberg A, Hunihan L, Ruddle F H. Proc Natl Acad Sci USA. 1994;91:6354–6358. doi: 10.1073/pnas.91.14.6354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim D, Ouyang H, Yang S H, Nussenzweig A, Burgman P, Li G C. J Biol Chem. 1995;270:15277–15284. doi: 10.1074/jbc.270.25.15277. [DOI] [PubMed] [Google Scholar]
- 39.DiCroce P A, Krontiris T G. Proc Natl Acad Sci USA. 1995;92:10137–10141. doi: 10.1073/pnas.92.22.10137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.May G, Sutton C, Gould H. J Biol Chem. 1991;266:3052–3059. [PubMed] [Google Scholar]
- 41.Hoff C M, Ghosh A K, Prabhakar B S, Jacob S T. Proc Natl Acad Sci USA. 1994;91:762–766. doi: 10.1073/pnas.91.2.762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Niu H, Zhang J, Jacob S T. Gene Expression. 1995;4:111–124. [PMC free article] [PubMed] [Google Scholar]
- 43.Niu H, Jacob S T. Proc Natl Acad Sci USA. 1994;91:9101–9105. doi: 10.1073/pnas.91.19.9101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Blier P R, Griffith A J, Craft J, Hardin J A. J Biol Chem. 1993;268:7594–7601. [PubMed] [Google Scholar]
- 45.Griffith A J, Blier P R, Mimori T, Hardin J A. J Biol Chem. 1992;267:331–338. [PubMed] [Google Scholar]
- 46.Mimori T, Hardin J A. J Biol Chem. 1986;261:10375–10379. [PubMed] [Google Scholar]
- 47.Li L L, Yeh N H. Exp Cell Res. 1992;199:262–268. doi: 10.1016/0014-4827(92)90433-9. [DOI] [PubMed] [Google Scholar]
- 48.Dvir A, Stein L Y, Calore B L, Dynan W S. J Biol Chem. 1993;268:10440–10447. [PubMed] [Google Scholar]
- 49.Mimori T, Ohosone Y, Hama N, Suwa A, Akizuki M, Homma M, Griffith A J, Hardin J A. Proc Natl Acad Sci USA. 1990;87:1777–1781. doi: 10.1073/pnas.87.5.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Griffith J D. Methods Cell Biol. 1973;7:129–145. doi: 10.1016/s0091-679x(08)61774-4. [DOI] [PubMed] [Google Scholar]
- 51.Griffith J D, Christiansen G. Annu Rev Biophys Bioeng. 1978;7:19–35. doi: 10.1146/annurev.bb.07.060178.000315. [DOI] [PubMed] [Google Scholar]
- 52.Arias J, Peterson S R, Dynan W S. J Biol Chem. 1991;266:8055–8061. [PubMed] [Google Scholar]
- 53.de Vries E, van Driel W, Bergsma W G, Arnberg A C, van der Vliet P C. J Mol Biol. 1989;208:65–78. doi: 10.1016/0022-2836(89)90088-0. [DOI] [PubMed] [Google Scholar]
- 54.Chan D W, Lees-Miller S P. J Biol Chem. 1996;271:8936–8941. doi: 10.1074/jbc.271.15.8936. [DOI] [PubMed] [Google Scholar]
- 55.Anderson C W. Trends Biochem Sci. 1993;18:433–437. doi: 10.1016/0968-0004(93)90144-c. [DOI] [PubMed] [Google Scholar]
- 56.Su W, Jackson S, Tjian R, Echols H. Genes Dev. 1991;5:820–826. doi: 10.1101/gad.5.5.820. [DOI] [PubMed] [Google Scholar]
- 57.Stenger J E, Tegtmeyer P, Mayr G A, Reed M, Wang Y, Wang P, Hough P V, Mastrangelo I A. EMBO J. 1994;13:6011–6020. doi: 10.1002/j.1460-2075.1994.tb06947.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mastrangelo I A, Courey A J, Wall J S, Jackson S P, Hough P V. Proc Natl Acad Sci USA. 1991;88:5670–5674. doi: 10.1073/pnas.88.13.5670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tuteja N, Tuteja R, Ochem A, Taneja P, Huang N W, Simoncsits A, Susic S, Rahman K, Marusic L, Chen J, Zhang J, Wang S, Ponger S, Falaschi A. EMBO J. 1994;13:4991–5001. doi: 10.1002/j.1460-2075.1994.tb06826.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Genersch E, Eckerskorn C, Lottspeich F, Herzog C, Kuhn K, Poschl E. EMBO J. 1995;14:791–800. doi: 10.1002/j.1460-2075.1995.tb07057.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Paillard S, Strauss F. Nucleic Acids Res. 1991;19:5619–5624. doi: 10.1093/nar/19.20.5619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rippe K, von Hippel P H, Langowski J. Trends Biochem Sci. 1995;20:500–506. doi: 10.1016/s0968-0004(00)89117-3. [DOI] [PubMed] [Google Scholar]
- 63.Schleif R. Annu Rev Biochem. 1992;61:199–223. doi: 10.1146/annurev.bi.61.070192.001215. [DOI] [PubMed] [Google Scholar]