Abstract
We have isolated and characterized a cDNA for a novel Per-Arnt/AhR-Sim basic helix–loop–helix (bHLH-PAS) factor that interacts with the Ah receptor nuclear translocator (Arnt), and its predicted amino acid sequence exhibits significant similarity to the hypoxia-inducible factor 1α (HIF1α) and Drosophila trachealess (dTrh) gene product. The HIF1α-like factor (HLF) encoded by the isolated cDNA bound the hypoxia-response element (HRE) found in enhancers of genes for erythropoietin, vascular endothelial growth factor (VEGF), and various glycolytic enzymes, and activated transcription of a reporter gene harboring the HRE. Although transcription-activating properties of HLF were very similar to those reported for HIF1α, their expression patterns were quite different between the two factors; HLF mRNA was most abundantly expressed in lung, followed by heart, liver, and other various organs under normoxic conditions, whereas HIF1α mRNA was ubiquitously expressed at much lower levels. In lung development around parturition, HLF mRNA expression was markedly enhanced, whereas that of HIF1α mRNA remained apparently unchanged at a much lower level. Moreover, HLF mRNA expression was closely correlated with that of VEGF mRNA. Whole mount in situ hybridization experiments demonstrated that HLF mRNA was expressed in vascular endothelial cells at the middle stages (9.5 and 10.5 days postcoitus) of mouse embryo development, where HIF1α mRNA was almost undetectable. The high expression level of HLF mRNA in the O2 delivery system of developing embryos and adult organs suggests that in a normoxic state, HLF regulates gene expression of VEGF, various glycolytic enzymes, and others driven by the HRE sequence, and may be involved in development of blood vessels and the tubular system of lung.
Keywords: vasculogenesis, tubular morphogenesis, trachealess
Vascular endothelial growth factor (VEGF) is a specific mitogen for endothelial cells that has been shown to be expressed in many tumor cell lines and vascular smooth muscle (1, 2) and is important not only for vasculogenesis and angiogenesis, but also for the maintenance of existing blood vessels (3). VEGF gene expression is induced by environmental stresses such as hypoxia, anemia, myocardial ischemia, and tumor progression to initiate subsequent angiogenesis and neovascularization (4–6).
Recent studies have revealed that the VEGF gene is induced through binding of a protein factor designated hypoxia-inducible factor 1 (HIF1) to the hypoxia response element (HRE) upstream of the VEGF gene (7–9). HIF1 was originally found as a critical mediator for inducible expression of the erythropoietin (Epo) gene by hypoxia (10), and has been shown to activate transcription of many genes including tyrosine hydroxylase (11), inducible nitric oxide synthase (12), and various glycolytic enzymes (13, 14) in a hypoxia-dependent manner. Recent cDNA cloning has demonstrated that HIF1 is a heterodimer composed of HIF1α and HIF1β (15). The latter is a factor already known as Ah receptor nuclear translocator (Arnt), which functions in association with the Ah receptor (AhR) as a mediator of various biological and toxicological effects of dioxin and other xenobiotics (16). HIF1α is a novel factor comprising a basic helix–loop–helix (bHLH) domain and a Per-Arnt/AhR-Sim (PAS) domain in the N terminus and exhibiting a striking homology with transcription factors encoded by Drosophila sim (dSim) (17), a master gene regulating the CNS midline development, and D. trachealess (dTrh) (18, 19), a regulator of development of the O2 delivery system such as trachea. Interestingly, a basic amino acid sequence immediately prior to the N terminus of the HLH domain, which is known to be the site for DNA recognition, is completely conserved between HIF1α and dTrh (18, 19).
In the course of searching for novel dimerization partners of Arnt, we isolated a cDNA clone encoding a bHLH-PAS protein closely related to HIF1α and dTrh, which we term HLF (HIF1α-like factor). Although HLF exhibits very similar characteristics to HIF1α in the properties of dimerization, DNA-binding, and transcriptional activation, their modes of expression are quite different from each other, in that HLF mRNA is abundantly expressed in a variety of organs including lung, heart, and other organs of adult mice in a normoxic state, whereas HIF1α mRNA is ubiquitous at much lower levels. In lung development around parturition, expression of HLF mRNA was markedly enhanced, while the low levels of HIF1α mRNA remained apparently unchanged. These expression patterns of HLF mRNA were found to closely parallel those of VEGF mRNA. Whole mount in situ hybridization experiments demonstrated that HLF was expressed in vascular endothelial cells at the middle stages of mouse embryo development where HIF1α was almost undetectable. Proposed regulatory functions of HLF include transcription of genes for VEGF and glycolytic enzymes and development of the vascular and pulmonary tubular system.
MATERIALS AND METHODS
Isolation of HLF cDNA.
The yeast two-hybrid screening was performed as described in the manufacturer’s protocol (Stratagene) using a prey phagemid library constructed from poly(A)-RNA isolated from ddy mouse hypothalamus with a cDNA insert for the bHLH-PAS domain of human Arnt as a bait plasmid (20). Approximately one million of yeast transformants were screened by β-galactosidase filter assay to isolate one positive clone. Screening a skeletal muscle cDNA library (21) with the isolated cDNA clone as a hybridization probe isolated two positive clones, and the cDNA inserts were sequenced in both directions.
Fluorescent in Situ Hybridization (FISH) for Chromosome Mapping.
Preparation of R-banded chromosomes and FISH were performed as described (22).
Recombinant Plasmids.
pEFBOS (23) and pGAD-Arnt (24) were described previously. pGBT-HIF1α (amino acids 1–324) and pGAD-HIF1α (amino acids 1–324) were constructed as follows. Oligonucleotide primers (5′-TCACCATGGAGGGCGCCGGC-3′ and 5′-TCATGTATCTTCTGATTCAACTTT-3′) were used to amplify the bHLH-PAS region of human HIF1α (hHIF1α), and the amplified products were inserted into the blunt-ended XmaI site of pGBT9 and pGAD424. pGBT-HLF and pGAD-HLF were constructed by inserting the SmaI cDNA fragment of HLF (amino acids 1–322) into the blunt-ended SalI sites of pGBT9 and pGAD424, respectively.
pSV40 promoter-EpoHRE-Luc plasmid was constructed by inserting the four HREs of the Epo gene (coding strand, 5′-GATCGCCCTACGTGCTGTCTCA-3′, the core sequence of the HRE is underlined) in tandem into the BglII site of pGL3 promoter plasmid (Promega). The pVEGF promoter (nucleotides 282-1230 in ref. 25)-Luc plasmid and pVEGF promoter (nucleotides 320-1230)-Luc plasmid were constructed by inserting the PCR-amplified DNAs into the SmaI site of pGL3 basic plasmid (Promega). The primers used for amplifications were 5′-TGCCAGACTACACAGTGCATACGTG-3′, 5′-AAGCCTCTGCGCTTCTCACC-3′, and 5′-GTCTCACTCCCCGCCACTGA-3′. pSV40 promoter-VegfHRE-Luc plasmid was constructed by inserting four copies of the HRE oligonucleotide (coding strand, 5′-TCGATACACAGTGCATACGTGGGTTTCCACAGGTCGTCT-3′, underlined as indicated above) of the VEGF gene into the SmaI site of the pGL3 promoter plasmid. pBOS-HIF1α, pBOS-HLF, and pBOS-Arnt were constructed by inserting full-length hHIF1α cDNA (a gift from I. Whitelaw and L. Poellinger, ref. 26), mouse HLF (mHLF) cDNA (nucleotides 1–3198), and hArnt (27), respectively, into the blunt-ended XbaI site.
Coimmunoprecipitation Assay.
Full-length cDNAs of HIF1α and HLF cloned in pBluescript vector were used to generate their 35S-labeled translation products in vitro by the TNT-coupled reticulocyte lysate system (Promega). Coimmunoprecipitation assay for protein interaction was performed as described (24).
Electrophoretic Mobility Shift Assay (EMSA).
The Epo and VEGF HRE double-stranded oligonucleotides were end-labeled by T4 DNA polynucleotide kinase with [γ-32P]ATP (ICN). A sequence of mutated Epo HRE (5′-GATCGCCCTAAAAGCTGTCTCA-3′, nucleotide substitutions are underlined) was used as negative control. The binding reactions were carried out as described previously (28).
Cell Lines and DNA Transfection.
A mutant cell line (c4) of Hepa-1 and Hep3B were maintained in DMEM supplemented with 10% fetal calf serum (FCS) in the presence and absence of 3.5% glucose, respectively. The expression plasmids were introduced into the cells by the Ca phosphate coprecipitation method together with a LacZ-expressing vector pENL as a standard for normalization as previously described (24).
Yeast Two-Hybrid System.
Protein–protein interactions were investigated by the two-hybrid system as described previously (24).
RNA Blot Analysis.
Total RNAs were prepared from various tissues of adult mice according to published protocols (29). Poly(A)-RNA was purified by using oligo(dT) cellulose (Sigma). Mouse HIF1α (mHIF1α) (nucleotides 2514–3414 in ref. 30), mHLF (nucleotides 1885–2685), mVEGF (nucleotides 50–951), and hG3PDH (CLONTECH) cDNAs were used as probes.
In Situ Hybridization.
Embryos at 9.5 days postcoitus (dpc) and 10.5 dpc were analyzed for HIF1α, HLF, and flt-1 expression by whole mount in situ hybridization with digoxygenin-labeled RNA used as probes, as described (31). mHIF1α probe (nucleotides 2514–3365), mHLF probe (nucleotides 3025–3488), and mflt-1 probe (a gift from M. Shibuya) in pBluescript were transcribed in the antisense and sense directions. In situ hybridization was performed essentially as described (31) except for a modification where hybridization was performed in the ExpressHyb solution (CLONTECH) containing 0.3–0.5 μg cRNA probe per ml at 65°C for 1 hr.
RESULTS
Cloning of mHLF, a Novel Member of Murine HIF1α Gene Family.
A novel cDNA clone was isolated from a murine hypothalamus cDNA library by a yeast two-hybrid system using the bHLH-PAS domain of Arnt as prey. Two additional independent clones were isolated as described in Materials and Methods to determine the entire coding sequence. The longest open reading frame encoded a novel polypeptide of 874 amino acids with a calculated molecular mass of 97 kDa. The sequence (ACAATGA) surrounding the putative initiation codon showed a reasonable agreement with the Kozak consensus sequence (5 out of 7; ref. 32). Sequence comparison in a protein database (GenBank) revealed that the amino acid sequence has a striking similarity to that of HIF1α in the amino-terminal half (amino acids 1–344) including bHLH (83.9% similarity) and PAS (66.5% similarity) motifs (Fig. 1A) followed by a sequence with a moderate similarity (36.4%, amino acids 345–559). While most of the sequence in the C-terminal half was variable between this protein and HIF1α, a small portion (63%, amino acids 824–874) of noticeable sequence similarity was found in the very C terminus. Thus, we designated this novel bHLH-PAS protein HLF. When the amino acid sequence in the bHLH-PAS region of HLF is compared with those of other members of the bHLH-PAS genes family, HLF is most closely related to HIF1α and dTrh (data not shown). It is notable that basic amino acids of the bHLH region involved in DNA recognition were completely conserved among HLF, HIF1α, and dTrh (Fig. 1B), suggesting that these factors recognize very similar, if not identical, regulatory DNA sequences.
Figure 1.
Comparison of primary structures of mouse HLF and HIF1α gene products. (A) Identical amino acid (∗) and conservative substitutions (.) are indicated. Dashes indicate deletions to maximize the sequence similarity. Basic helix–loop–helix region and direct repeats of PAS domain are boxed. The entire PAS domain is enclosed in brackets. (B) Comparison of the basic amino acid sequence of Arnt (33), dSim (17), AhR (34, 35), HIF1α (15), HLF, and dTrh (18, 19). Amino acids identical to those of HLF are boxed.
Chromosomal Localization of mHLF.
FISH experiments using HLF cDNA as probe were performed to determine chromosomal localization of the HLF gene. Hybridization signals were detected as twin spots at the E4–E5 band of mouse chromosome 17 (Fig. 2). Because the HIF1α gene is reported to be localized on mouse chromosome 12 (36), members of the HIF1α gene family are not clustered in the mouse genome.
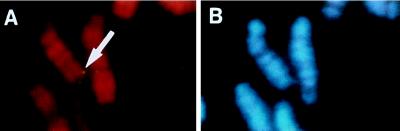
Figure 2.
Chromosomal localization of HLF gene on mouse R-banded chromosomes. The hybridization signals are indicated by an arrow. The metaphase spreads were hybridized as described (22) and photographed with Nikon B-2A (A) and UV-2A (B) filters. R-banded and G-banded patterns are demonstrated in (A) and (B), respectively.
Dimerization, DNA Binding, and Transcription-Activating Properties of HLF.
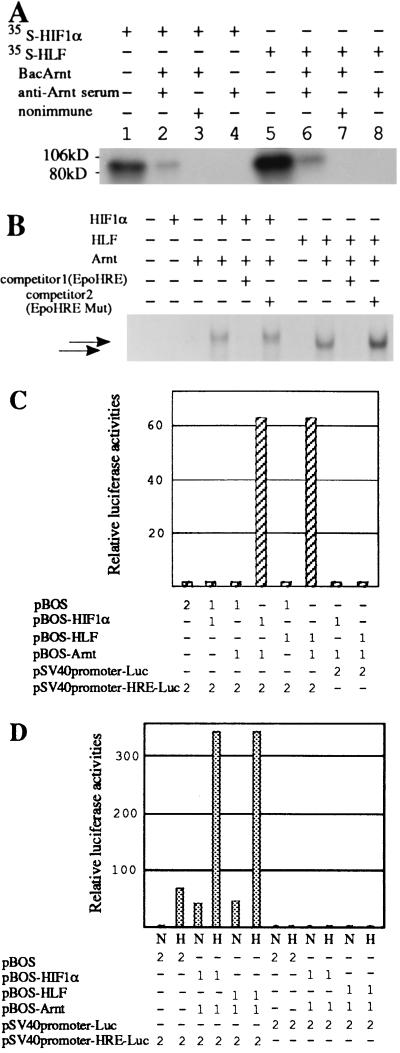
HIF1α forms a dimer with Arnt and regulates hypoxia-inducible Epo expression through binding to the 3′-hypoxia response enhancer (15). To clarify whether HLF forms a functional heterodimer with Arnt in a similar manner to HIF1α, we investigated the mode of HLF and HIF1α dimerization with Arnt by coimmunoprecipitation assays and the yeast two-hybrid system. As shown in Table 1, both HLF and HIF1α were found to interact selectively with Arnt in yeast cells. No interaction between HIF1α and HLF was found. In agreement with these results, HIF1α and HLF were coimmunoprecipitated by anti-Arnt antibodies, only when they were incubated with Arnt (Fig. 3A). EMSA experiments using the Epo HRE sequence showed that the HLF/Arnt heterodimer recognized and bound the HRE with similar intensity to the HIF1α/Arnt heterodimer (Fig. 3B). DNA transfer experiments using the HIF1α, HLF, and Arnt expression plasmids demonstrated that the combined expressions of HLF and Arnt synergistically activated transcription under the control of tandem arrays of the Epo HRE, in similar fashion to the heterodimer of HIF1α and Arnt (Fig. 3C). Maintenance of the cells in a hypoxic condition further enhanced the activated transcription from the reporter plasmid during coexpression either of HLF and Arnt, or of HIF1α and Arnt (Fig. 3D). Hypoxic enhancement of the transcription driven by the Epo HRE has already been reported with coexpression of HIF1α and Arnt (8). Taken together, these results show that HLF has biochemical- and transcription-activating properties very similar to HIF1α.
Table 1.
Interaction between HLF and Arnt as revealed by the yeast two-hybrid system
| GAL-DBD fusions | GAL activation domain fusions
|
|||
|---|---|---|---|---|
| GAD424 | Arnt | HIF1α | HLF | |
| pGBT9 | <0.02* | <0.02 | <0.02 | <0.02 |
| Arnt | < 0.02 | 0.066 ± 0.016 | 1.71 ± 0.24 | 1.61 ± 0.1 |
| HIF1α | < 0.02 | 14.5 ± 5.9 | < 0.02 | < 0.02 |
| HLF | < 0.02 | 13.2 ± 3.0 | < 0.02 | < 0.02 |
Yeast strain SFY526 containing lacZ under the control of the GAL1 promoter was cotransfected with the indicated plasmids, and β-galactosidase activities were determined.
β-galactosidase activity (Miller units).
Figure 3.
Coimmunoprecipitation, EMSA, and transient transfection experiments. (A) Interaction of HLF or HIF1α with Arnt was investigated by coimmunoprecipitation assay using extracts from Sf9 cells transfected with baculovirus-carrying Arnt cDNA and 35S-labeled HLF or HIF1α proteins synthesized in the in vitro reticulocyte translation system as described. Lanes 1 and 5, input proteins. (B) EMSA experiment using the Epo HRE oligonucleotide. Combinations of the reaction components are indicated above the blot. (C) Transient transfection experiment in c4 cells. Combinations of effector and reporter plasmids and their amounts (μg) are indicated beneath the chart. (D) Effects of hypoxic treatment on the transcription of the reporter gene in Hep3B cells. Combinations of effector and reporter plasmids used for transfection and their amounts (μg) are indicated below the chart. Cells were grown under normoxic (N, 21% O2) and hypoxic (H, 1% O2) conditions.
Tissue Distribution of HIF1α and HLF mRNA.
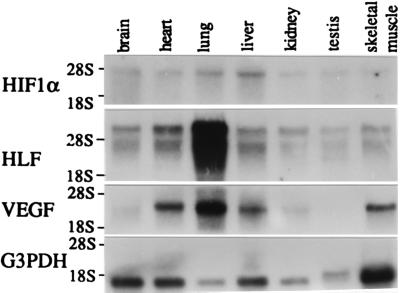
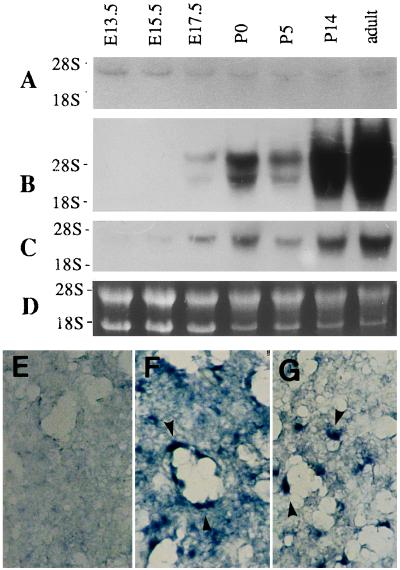
To further study functional roles of HLF, the tissue distributions of the HLF and HIF1α mRNAs were investigated by RNA blot analysis. mRNAs for HLF and HIF1α were expressed more or less ubiquitously in all tissues examined (Fig. 4). The level of HLF mRNA expression, however, was generally much higher (at least 10-fold) than that of HIF1α mRNA, because the time for exposure in autoradiography was one order of the magnitude longer with HIF1α (2 weeks) than with HLF (36 hr) in Fig. 4. Interestingly, the VEGF gene, which is known to be transcriptionally activated by HIF1 (7–9), was also expressed abundantly in lung, heart, and liver, and was hardly detectable in testis. Its expression patterns appeared to be coordinated with that of HLF rather than that of HIF1α. To pursue this relationship between HLF and VEGF mRNA expression patterns, expression of HLF and VEGF during lung development was examined by RNA blot analysis (Fig. 5). HLF and VEGF mRNAs were expressed scarcely in the lung of 13.5 dpc and 15.5 dpc, and became prominent in the 17.5-dpc and postnatal day 0 (P0) lung, and highly abundant in the adult lung. On the other hand, HIF1α expression levels were constantly low throughout lung development. The detailed expression of HLF and VEGF mRNAs in the lung was determined by in situ hybridization experiments. VEGF mRNA was expressed in the epithelial cells of pulmonary alveoli in the P0 and adult lung. HLF mRNA was coexpressed with VEGF mRNA ubiquitously and intensely at least in alveolar epithelial cells of the P0 (Fig. 5) and adult mice (data not shown), whereas HIF1α was not significantly expressed in these cells.
Figure 4.
RNA blot analysis using HIF1α and HLF cDNA probes. poly(A)-RNAs (3 μg) prepared from various tissues of mice, as described, were electrophoresed in a 0.8% agarose gel containing 2.2 M formaldehyde and then transferred to a nylon membrane. The membrane was probed with 32P-labeled mHIF1α cDNA (A), mHLF cDNA (B), mVEGF (C), and hG3PDH (D). Tissues from which poly(A)-RNAs were prepared are indicated across the top.
Figure 5.
mRNA expression of HIF1α, HLF, and VEGF in the process of lung development. (A–C) RNA blot analysis was performed by using total RNAs (15 μg) from the lungs of embryos of 13.5 dpc (E13.5 dpc) to adult mouse with probes for HIF1α (A), HLF (B), and VEGF (C). (D) Ethidium bromide-staining of RNA electrophoresed on agarose gel. (E–G) In situ hybridization analysis of mouse lung of the postnatal day 0 (P0) by the cRNA probes for HIF1α (E), HLF (F), and VEGF (G). Coloring reaction was done for 1 day for F and 3 days for E and G. Arrowheads indicate the expression of alveolar epithelial cells.
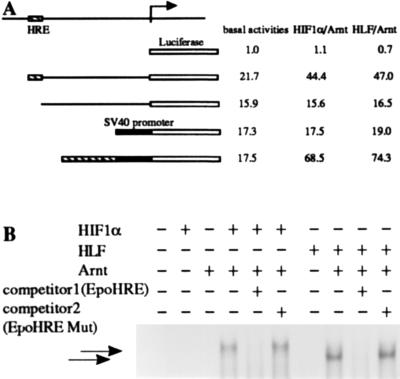
Transcriptional Regulation of the VEGF Gene by the HLF/Arnt Heterodimer.
The coordinated expression of HLF and VEGF suggested that HLF may regulate the VEGF gene during the developmental process and in adult tissues. Thus, we investigated whether HLF activates the transcription of the VEGF gene through binding to the HRE sequence upstream of the VEGF gene. The VEGF promoter region (nucleotides 282-1230) was ligated to the upstream of the firefly luciferase gene (Fig. 6A), and the constructed reporter plasmid was cotransfected into c4 cells with a pair of expression plasmids for either HLF and Arnt, or for HIF1α and Arnt. The transcriptional activation of the VEGF (nucleotides 282-1230)-Luc reporter plasmid by HIF1α/Arnt or HLF/Arnt heterodimer was activated 2.5-fold over that of the VEGF reporter plasmid lacking the HRE. Deletion of the HRE sequence located 0.9 kb upstream of the transcription initiation site completely abolished transcriptional activation by either HIF1α/Arnt or HLF/Arnt. The two heterodimers activated transcription of reporter plasmid under the control of tandemly repeated HREs by 5-fold. EMSA experiments in Fig. 6B using the HRE oligonucleotide of the VEGF promoter showed that in vitro translated HLF recognized and bound the HRE of the VEGF gene in a complex with Arnt, in similar fashion to HIF1α. Formation of these complexes was competed out with an excess amount of the Epo HRE sequence. From these results, together with those of the preceding section, it is reasonable to consider that HLF rather than HIF1α regulates transcription of the VEGF gene during developmental processes and in adult tissues under normoxic conditions.
Figure 6.
Transcriptional regulation of the VEGF gene by a heterodimer of HLF and Arnt. (A) Schematic representation of the VEGF gene promoter region and reporter gene, and summary of the transfection assay. The VEGF gene promoter and the HRE-deleted promoter regions were PCR-amplified and ligated to the firefly luciferase reporter gene as described. The VEGF HRE sequences were tandemly ligated to the promoter of the heterologous SV40 promoter. Plasmid DNAs of a reporter (2 μg), effectors (1 μg), and β-galactosidase expressing pENL (1 μg) were cotransfected, and expressed luciferase and β-galactosidase activities were determined. (B) EMSA experiment using the HRE oligonucleotide of the VEGF gene. Combinations of various proteins components and competitor DNA are indicated above the blot.
Spatiotemporal Expression of HIF1α and HLF in Early Embryogenesis.
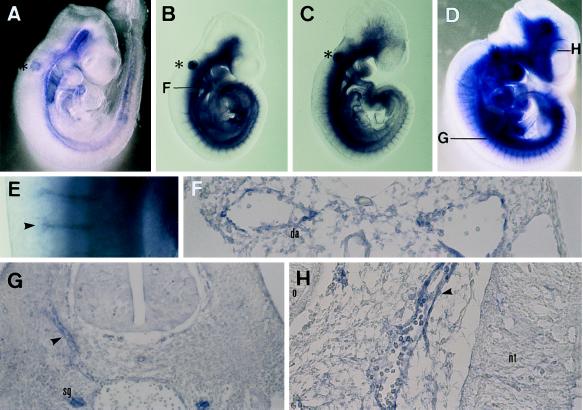
To gain an insight into their possible functions in mouse embryogenesis, spatiotemporal distribution patterns of HIF1α and HLF mRNAs were investigated by whole-mount in situ hybridization at 9.5 and 10.5 dpc. HLF mRNA was expressed abundantly in the vascular system in 9.5-dpc embryos (Fig. 7 B and D–H), and a transverse section showed that HLF was expressed in the endothelial cells of the dorsal aorta (Fig. 7F). HIF1α was also expressed in similar regions to HLF, albeit at much lower levels (Fig. 7A). Expression in the vascular endothelial cells was confirmed by the expression of flt-1 (Fig. 7D) and flk1 (data not shown) used as reference standards for vascular endothelial cells expression (37, 38). In 10.5-dpc embryos, HLF became prominent in the sympathetic ganglion and sprouting vessels of the head mesenchyme (Fig. 7 G and H), in addition to high expression in vascular endothelial cells, including the dorsal aorta and intersomitic plexus, whereas HIF1α expression remained very low in the same tissue as it was at 9.5 dpc (data not shown).
Figure 7.
Whole-mount in situ hybridization on mouse embryos using HLF, HIF1α, and flt-1 cRNA probe. (A) Lateral view of 9.5-dpc embryo hybridized with HIF1α cRNA probe. Hybridization signal was seen weakly in the vascular system. ∗, nonspecific signal in the otic vesicle. (B) Lateral view of 9.5-dpc embryo hybridized with HLF cRNA probe. Hybridization signal was observed clearly in the vascular system. F, position of cross-section in F. (C) Lateral view of 9.5-dpc embryo hybridized with flt-1 cRNA probe. (D) Lateral view of 10.5-dpc embryo hybridized with HLF cRNA probe. E, photograph of higher magnification (×45) in E. G and H, position of the cross-section in G and H. (E) Photograph of higher magnification (×45) of dorsal region of 10.5-dpc embryo. Arrows indicate the signals in the intersomitic plexus. (F) Transverse section of 9.5-dpc embryo hybridized with HLF cRNA. Note that HLF mRNA was expressed in the endothelial cells of the dorsal aorta. (G) Transverse section of 10.5-dpc embryo. Arrowheads indicate the signals in the intersomitic plexus. (H) Transverse section of the 10.5-dpc embryo. An arrowhead indicates the signal of the endothelial cells of the sprouting blood vessels. da, dorsal aorta: nt, neural tube: o, optic vesicle; sg, sympathetic ganglion.
DISCUSSION
We have described isolation of cDNA for a novel bHLH-PAS protein closely related to HIF1α, designated HLF. Comparison of its amino acid sequence with those of other bHLH-PAS proteins reported so far revealed that the sequence of the bHLH-PAS domain sequence is highly conserved among HLF, HIF1α, and dTrh. In particular, the basic sequences immediately prior to the N terminus of the HLH domains, which are directly involved in the DNA binding, show a perfect identity with one another (Fig. 1B). This suggests that these factors recognize very similar, if not identical, DNA regulatory sequences and may regulate a similar group of genes. This suggestion was substantiated by the coimmunoprecipitation and transient DNA transfection experiments in which HLF clearly formed a dimer with Arnt and activated reporter gene transcription driven by either VEGF or Epo HRE. The EMSA experiments also demonstrated that both HLF and HIF1α bound the VEGF and Epo HRE sequences in association with Arnt, and with a similar affinity.
In contrast to their very similar biochemical properties, the HLF and HIF1α mRNAs showed distinct modes of expressions, as revealed by RNA blot analysis and in situ hybridization. Although the two mRNAs appeared to be ubiquitously expressed in all the tissues examined, the level of expression markedly differed between HLF and HIF1α in various tissues. HLF mRNA was expressed most abundantly in the lung, followed by heart and liver of adult mice, whereas HIF1α mRNA was expressed at a much reduced level (less than 10-fold). Interestingly, variation in the HLF mRNA content in various tissues closely paralleled that of the VEGF mRNA. This tendency appeared to be more prominent during development of the lung. Expression of VEGF mRNA in the lung was markedly increased from the late fetal stage in parallel with enhanced expression of HLF mRNA, whereas HIF1α mRNA remained constant at a much lower level. Furthermore, in situ hybridization experiments revealed that the VEGF mRNA was expressed in alveolar epithelial cells, where HLF mRNA was always coexpressed. Taken together with the structural similarity between HLF and dTrh, and the observation that HIF1α was expressed at a much reduced level in these cells (Fig. 5 A and E), these results strongly suggest that HLF regulates the expression of the VEGF gene in a normoxic state through binding to the HRE sequence and is involved in tubular morphogenesis of lung and blood vessels. In a hypoxic state, HIF1α is known to be involved in inducible expression of the Epo gene in the Hep3B human hepatoma cell line (15).
In situ hybridization experiments with 9.5-dpc, 10.5-dpc (Fig. 7), and 13.5-dpc embryos (data not shown) showed that HLF mRNA was expressed in the endothelial cells of the intersomitic plexus and dorsal aorta with an expression pattern very similar to that of flk-1 and flt-1, which are known to be expressed in the vascular endothelial cells (37, 38). In contrast, HIF1α was expressed at such low levels at these developmental stages that its precise expression could not be localized. At those stages of embryonic development, the VEGF gene is reported to be expressed in smooth muscle cells lining the vascular endothelium and to function as a growth factor in angiogenesis and vasculogenesis (39), whereas flt-1 and flk-1 function as receptors for VEGF in the vascular endothelial cells in a paracrine manner. Because of a very low level of HLF mRNA expression in the smooth muscle cells of the vascular system, it remains to be clarified whether HLF was coexpressed with VEGF in vascular smooth muscle cells at those stages of vascular development. Because it is considered to regulate potentially many genes such as those for PDGF-B (40), endothelin-1 (33), and various glycolytic enzymes (13, 14) by binding to the HRE, HLF may play physiological roles other than the proposed regulation of VEGF mRNA expression in vasculogenesis and maintenance of blood vessels.
Acknowledgments
The authors thank Drs. M. Shibuya (The University of Tokyo, Tokyo) for mflt-1 and mflk-1 cDNA, L. Poellinger (Karolinska Institute, Stockholm) for hHIF1α cDNA, M. Gassmann (University of Zurich–Irchel, Zurich) for mHIF1α cDNA, M. Whitelaw (The University of Adelaide, Adelaide) for critically reading this manuscript, and Mr. H. Abe (The University of Tohoku, Sendai) for help in transfections experiments. This work was supported in part by Grants-in-Aid for Scientific Research on Priority Areas, for Scientific Research (B); and the International Scientific Research Program from the Ministry of Education, Culture, Sport and Science of Japan; and by a fund from Sankyo. M.E. is a research fellow of the Japan Society for the Promotion of Science.
ABBREVIATIONS
- VEGF
vascular endothelial growth factor
- Epo
erythropoietin
- AhR
Ah receptor
- HIF1α
hypoxia-inducible factor
- P0
postnatal day 0
- EMSA
electrophoretic mobility shift assay
- HLF
HIF1α-like factor
- HRE
hypoxia-response element
- Arnt
Ah receptor nuclear translocator
- dTrh, Drosophila trachealess
dpc, days postcoital
- bHLH
basic helix–loop–helix
- PAS
Per-Arnt/AhR-Sim
Footnotes
References
- 1.Ferrara N, Houck K, Jakeman L, Leung D. Endocr Rev. 1992;13:18–32. doi: 10.1210/edrv-13-1-18. [DOI] [PubMed] [Google Scholar]
- 2.Dvorak H F, Brown L F, Detmar M, Dvorak A M. Am J Pathol. 1995;146:1029–1039. [PMC free article] [PubMed] [Google Scholar]
- 3.Berse B, Brown L F, Van De Water, Dvorak H F, Senger D R. Mol Biol Cell. 1992;3:211–220. doi: 10.1091/mbc.3.2.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Banai S, Shweiki A, Pinson A, Chandra M, Lazarovici G, Keshet E. Cardiovasc Res. 1994;28:1176–1179. doi: 10.1093/cvr/28.8.1176. [DOI] [PubMed] [Google Scholar]
- 5.Plate K H, Breier G, Weich A, Risau W. Nature (London) 1992;359:845–848. doi: 10.1038/359845a0. [DOI] [PubMed] [Google Scholar]
- 6.Shweiki D, Itin A, Soffer D, Keshet E. Nature (London) 1992;359:843–845. doi: 10.1038/359843a0. [DOI] [PubMed] [Google Scholar]
- 7.Levy A P, Levy N S, Wegner S, Goldberg M A. J Biol Chem. 1995;270:1333–13340. doi: 10.1074/jbc.270.22.13333. [DOI] [PubMed] [Google Scholar]
- 8.Forsythe J A, Jiang B-H, Iyer N V, Agani F, Leung S W, Koos R D, Semenza G L. Mol Cell Biol. 1996;16:4604–4613. doi: 10.1128/mcb.16.9.4604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu Y, Cox S R, Morita T, Kourembanas S. Circ Res. 1996;77:638–643. doi: 10.1161/01.res.77.3.638. [DOI] [PubMed] [Google Scholar]
- 10.Semenza G L, Wang G L. Mol Cell Biol. 1992;12:5447–5454. doi: 10.1128/mcb.12.12.5447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Norris M L, Millhorn D E. J Biol Chem. 1996;270:23774–23779. doi: 10.1074/jbc.270.40.23774. [DOI] [PubMed] [Google Scholar]
- 12.Melillo G, Musso T, Sica A, Taylor L S, Cox G W, Varesio L. J Exp Med. 1995;182:1683–1693. doi: 10.1084/jem.182.6.1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Firth J D, Ebert B L, Pugh C W, Ratcliffe P J. Proc Natl Acad Sci USA. 1994;91:6496–6500. doi: 10.1073/pnas.91.14.6496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Semenza G L, Roth P H, Fang H M, Wang G L. J Biol Chem. 1995;269:23757–23763. [PubMed] [Google Scholar]
- 15.Wang G L, Jiang B-H, Rue E A, Semenza G L. Proc Natl Acad Sci USA. 1995;92:5510–5514. doi: 10.1073/pnas.92.12.5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hankinson O. Annu Rev Pharmacol Toxicol. 1995;35:307–340. doi: 10.1146/annurev.pa.35.040195.001515. [DOI] [PubMed] [Google Scholar]
- 17.Nambu J R, Lewis J O, Wharton K O, Jr, Crews S T. Cell. 1991;67:1157–1167. doi: 10.1016/0092-8674(91)90292-7. [DOI] [PubMed] [Google Scholar]
- 18.Isaac D D, Andrew D J. Genes Dev. 1996;10:103–118. doi: 10.1101/gad.10.1.103. [DOI] [PubMed] [Google Scholar]
- 19.Wilk R, Weizman I, Shilo B-Z. Genes Dev. 1996;10:93–102. doi: 10.1101/gad.10.1.93. [DOI] [PubMed] [Google Scholar]
- 20.Hirose K, Morita M, Ema M, Mimura J, Hamada H, Fujii H, Saijoh Y, Gotoh O, Sogawa K, Fujii-Kuriyama Y. Mol Cell Biol. 1996;16:1706–1713. doi: 10.1128/mcb.16.4.1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ema M, Suzuki M, Morita M, Hirose K, Sogawa K, Matsuda Y, Gotoh O, Saijoh Y, Fujii H, Hamada H, Fujii-Kuriyama Y. Biochem Biophys Res Commun. 1996;218:588–594. doi: 10.1006/bbrc.1996.0104. [DOI] [PubMed] [Google Scholar]
- 22.Matsuda Y, Harada Y-H, Natsume-Sakai S, Lee K, Shiomi T, Chapman V M. Cytogenet Cell Genet. 1992;61:282–285. doi: 10.1159/000133423. [DOI] [PubMed] [Google Scholar]
- 23.Mizushima S, Nagata S. Nucleic Acids Res. 1990;18:5322. doi: 10.1093/nar/18.17.5322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ema M, Morita M, Ikawa S, Tanaka M, Matsuda Y, Gotoh O, Saijoh Y, Fujii H, Hamada H, Kikuchi Y, Fujii-Kuriyama Y. Mol Cell Biol. 1996;16:5865–5875. doi: 10.1128/mcb.16.10.5865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shima D T, Kuroki M, Deutsch U, Ng Y-S, Adamis A P, D’Amore P A. J Biol Chem. 1996;271:3877–3883. doi: 10.1074/jbc.271.7.3877. [DOI] [PubMed] [Google Scholar]
- 26.Gradin K, McGuire J, Wenger R H, Kvietikova I, Whitelaw M L, Toftgard R, Tora L, Gassmann M, Poellinger L. Mol Cell Biol. 1996;16:5221–5231. doi: 10.1128/mcb.16.10.5221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hoffman E C, Reyes H, Chu F-F, Sander F, Conley L H, Brooks B H, Hankinson O. Science. 1991;252:954–958. doi: 10.1126/science.1852076. [DOI] [PubMed] [Google Scholar]
- 28.Sogawa K, Nakano R, Kobayashi A, Kikuchi Y, Ohe N, Matsushita N, Fujii-Kuriyama Y. Proc Natl Acad Sci USA. 1995;92:1936–1940. doi: 10.1073/pnas.92.6.1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chirgwin J M, Przybyla A E, MacDonald R J, Rutter W J. Biochemistry. 1979;18:5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- 30.Wenger R H, Rolfs A, Marti H H, Guenet J L, Gassmann M. Biochem Biophys Res Commun. 1996;223:54–59. doi: 10.1006/bbrc.1996.0845. [DOI] [PubMed] [Google Scholar]
- 31.Wilkinson D G. In Situ Hybridization: A Practical Approach. London: Oxford Univ. Press; 1992. [Google Scholar]
- 32.Kozak M. J Cell Biol. 1989;108:229–241. doi: 10.1083/jcb.108.2.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kourembanas S, Marsden P A, McQuillan L P, Faller D V. J Clin Invest. 1991;88:1054–1057. doi: 10.1172/JCI115367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ema M, Sogawa K, Watanabe N, Chujoh Y, Matsushita N, Gotoh O, Funae Y, Fujii-Kuriyama Y. Biochem Biophys Res Commun. 1992;184:246–253. doi: 10.1016/0006-291x(92)91185-s. [DOI] [PubMed] [Google Scholar]
- 35.Burbach K, Poland M A, Bradfield C A. Proc Natl Acad Sci USA. 1992;89:8185–8189. doi: 10.1073/pnas.89.17.8185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Semenza G L, Rue E A, Iyer N V, Pang M G, Kearns W G. Genomics. 1996;34:437–439. doi: 10.1006/geno.1996.0311. [DOI] [PubMed] [Google Scholar]
- 37.Breier G, Clauss M, Risau W. Dev Dyn. 1995;204:228–239. doi: 10.1002/aja.1002040303. [DOI] [PubMed] [Google Scholar]
- 38.Millauer B, Wizigmann-Voos S, Schnurch H, Martinez R, Moler N P H, Risau W, Ullrich A. Cell. 1993;72:835–846. doi: 10.1016/0092-8674(93)90573-9. [DOI] [PubMed] [Google Scholar]
- 39.Peters K G, De Vries C, Williams L T. Proc Natl Acad Sci USA. 1993;90:8915–8919. doi: 10.1073/pnas.90.19.8915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kourembanas S, Hannan R L, Faller D V. J Clin Invest. 1990;86:670–674. doi: 10.1172/JCI114759. [DOI] [PMC free article] [PubMed] [Google Scholar]