Abstract
GDP-dissociation inhibitors (GDIs) play a primary role in modulating the activation of GTPases and may also be critical for the cellular compartmentalization of GTPases. RhoGDI and GDI/D4 are two currently known GDIs for the Rho-subfamily of GTPases. Using their cDNAs to screen a human brain cDNA library under low stringency, we have cloned a homologous cDNA preferentially expressed at high levels in brain and pancreas. The predicted protein, named RhoGDIγ, is ≈50% identical to GDI/D4 and RhoGDI. It binds to CDC42 and RhoA with less affinity compared with RhoGDI and does not bind with Rac1, Rac2, or Ras. RhoGDIγ functions as a GDI for CDC42 but with ≈20 times less efficiency than RhoGDI. Immunohistochemical studies showed a diffuse punctate distribution of the protein in the cytoplasm with concentration around the nucleus in cytoplasmic vesicles. Overexpression of the protein in baby hamster kidney cells caused the cells to round up with loss of stress fibers. A distinct hydrophobic amino terminus in RhoGDIγ, not seen in the other two RhoGDIs, could provide a mechanism for localization of the GDI to specific membranous compartment thus determining function distinct from RhoGDI or GDI/D4. Our results provide evidence that there is a family of GDIs for the Rho-related GTPases and that they differ in binding affinity, target specificity, and tissue expression. We propose that RhoGDI be renamed RhoGDIα and GDID4 be renamed RhoGDIβ. The new GDI should widen the scope of investigation of this important class of regulatory protein.
A key biochemical activity of all small GTP-binding proteins is their cycling between a GDP-bound inactive state and the GTP-bound active state. Three classes of proteins are currently known to regulate this critical switching of molecular states (1). The GDP-dissociation stimulators or GDP exchange factors catalyze the exchange of GDP for GTP, the GTPase activating proteins catalyze the intrinsic ability of the GTP-binding proteins to hydrolyze GTP to GDP, and the GDP-dissociation inhibitors (GDIs) inhibit the exchange of GDP for GTP.
An array of exchange factors have thus far been isolated for the Rho family (RhoA, B, C, G, Rac1, Rac2, and CDC42). They all have in common a dbl-homology domain responsible for stimulating nucleotide exchange activity. In contrast, only two GDIs for the Rho-related proteins have been identified so far. A RhoGDI protein was first isolated and cloned from bovine brain cytosol (2). Leonard et al. (3), in searching for a GDI for CDC42Hs, isolated a GDI from bovine brain cytosol that is virtually identical to the bovine RhoGDI. Subsequently RhoGDI was found to be capable of functioning also as a GDI for Rac1 (4) and Rac2 (5).
By subtractive hybridization we cloned a RhoGDI-homologous cDNA, named GDI/D4 (6), representing an mRNA that is expressed at a very high level only in hematopoietic cells. GDI/D4 is 67% identical to RhoGDI. In vitro studies showed that GDI/D4 also can function as a GDI for Rho, Rac, and CDC42 (7, 8).
The in vitro biochemical activity of these GDIs suggests that their role may be to inhibit the generation of active GTP-bound Rho proteins. Thus microinjection of RhoGDI into fibroblasts caused inhibition of motility (9) and overexpression of RhoGDI and GDI/D4 in various cell lines induced disruption of the actin cytoskeleton and the rounding up of cells (10, 11). In contrast, RhoGDI has also been shown to be an inhibitor of the intrinsic and GTPase activating protein-stimulated GTP hydrolytic activity of CDC42 (12), Rac (13), and Rho (14), and thus possesses the ability to maintain these proteins in the GTP-bound active form. Thus RhoGDI appears to be a molecule capable of blocking the GTP-binding/GTPase cycle at two points—i.e., at the GDP–GTP exchange step and at the GTP hydrolytic step.
Although both GDI/D4 and RhoGDI function as GDIs for the same spectrum of substrates, GDI/D4 has been shown to bind to the Rho subfamily of proteins with a significantly lower affinity compared with RhoGDI. A replacement of residues 169–178 of GDI/D4 with the homologous region from RhoGDI converted its activity to resemble that of RhoGDI (15). These data suggest that GDI/D4 and RhoGDI very likely have overlapping function. While macrophages with loss of function of GDI/D4 showed an impairment in their capacity to generate superoxide (16), the absence of a more severe phenotypic effect is likely due to a redundancy of function between the two GDIs. This raises the issue of whether other GDIs, homologous to RhoGDI and GDI/D4, exist. We therefore screened cDNA libraries under low stringency with RhoGDI and GDI/D4 to identify crosshybridizing clones. A homologous cDNA was identified, which we named RhoGDIγ.
MATERIALS AND METHODS
Cloning of cDNA.
Duplicate filters of a phage cDNA library in the λ gt11 vector (CLONTECH) derived from human fetal brain RNAs were screened with the human GDI/D4 and RhoGDI cDNAs as described (17). Hybridization and washing were first carried out under high-stringency conditions (65°C, 6× standard saline citrate). Positive clones representing clones for GDI/D4 or RhoGDI were identified by autoradiography. A triplicate filter was then hybridized under low stringency (50°C, 6× standard saline citrate) and newly hybridizing clones, representing clones that crosshybridize with GDI/D4 or RhoGDI, were identified. Individual clones were then isolated after two rounds of secondary screening under similar low stringency condition.
Expression Studies.
Northern blot filters of poly(A)+ RNA from normal human tissues were purchased from CLONTECH. The filters were probed with 32P-labeled cDNA under standard conditions (17).
Construction of Expression Vectors and Production of Fusion Protein.
Using PCR, cDNA fragments with the restriction sites EcoRI and SalI introduced into the 5′ and 3′ ends respectively of the ORF for RhoGDIγ were generated. The PCR product was ligated directionally into the EcoRI and SalI sites of the pGEX4T-1 vector as described (15). In a similar maneuver, a truncated cDNA was generated and ligated into the pGEX4T-1 vector to produce a glutathione S-transferase (GST) fusion protein with the first 30 amino acids of RhoGDIγ protein deleted. GST–GDI plasmids were transformed into bacteria and GST–GDI fusion proteins isolated as described (3).
Biochemical Function Studies.
Baculovirus for CDC42Hs and RhoA (obtained from Christopher Carpenter, Harvard Medical School, Boston), and for Rac1 and Rac2 (obtained from Gary Bokoch, Scripps Research Institute, La Jolla, CA) were used to infect Spodoptera frugiperda (Sf9) insect cells and harvested after 24–48 hr. Cell pellets were rinsed, resuspended in lysing buffer, freeze-thawed several times, and lysate-clarified by high-speed centrifuge. The lysates were incubated with GST-RhoGDIγ to test for binding. The capacity of RhoGDIγ to inhibit the dissociation of GDP from GTP-binding proteins was measured by the nitrocellulose filter binding assay using CDC42Hs produced in Sf9 cells as described (3),
Green Fluorescent Protein (GFP)-Fusion Protein Vector.
A PCR cDNA fragment containing the entire coding region of RhoGDIγ was generated (5′ primer, 5′-GCCGCGGTCGACATGCTGGGCCTGGACGCGTGCGAGCTG-3′; 3′ primer, 5′-AACTGAGAATTCTCAGGGGAGACACGGACTGGG-3′) that allowed cloning of the cDNA into the SalI (5′) and BamHI (3′) sites of the pEGFP-C1 vector (CLONTECH). The vector generates a GFP-tagged RhoGDIγ protein with the GFP fused in-frame to the amino-terminal end of the protein. The DNA was transfected into baby hamster kidney cells as described using the calcium phosphate precipitation technique and after various time intervals the cells were fixed and examined under argon arc light activation to determine the cellular location of GFP-tagged proteins.
DNA Sequencing.
DNA sequencing was done using the Applied Biosystems model 373A automated DNA sequencer and the Taq dideoxy terminator cycle sequencing method.
RESULTS
Isolation and Molecular Analysis of RhoGDIγ cDNA.
A total of one million plaques from a human brain cDNA library were screened with the human GDI/D4 and human RhoGDI cDNA probes under high stringency. Positive clones were identified and marked. The filters were then probed with GDI/D4 and RhoGDI under low stringency. Additional hybridizing clones were identified. Ten clones were randomly selected for secondary screening to obtain individual hybridizing clones. These were sequenced in single runs to obtain nucleotide sequence of at least 250 nucleotides for every clone. Two clones were identified that showed homology to RhoGDI and GDI/D4. One of them was then used to screen the brain cDNA library again under high stringency to obtain additional clones. A clone with the longest cDNA insert of ≈1.2 kb was isolated and sequenced to completion. This clone was named RhoGDIγ.
Fig. 1 shows the nucleotide sequence of the human cDNA. There is a long open reading frame from nucleotide 1 to a stop codon TGA at nucleotide 714 with the first methionine at nucleotide 39. There is no stop codon upstream of the first methionine. The predicted amino acid sequence from the ORF revealed a protein of 225 amino acids.
Figure 1.
Nucleotide and predicted amino acid sequence of RhoGDIγ. Nucleotides are numbered together with amino acid sequence of the longest ORF numbered from the presumed initiating methionine. A TGA stop codon (∗∗∗) is followed by a short 3′ untranslated region.
Fig. 2 shows that RhoGDIγ is homologous to RhoGDI and GDI/D4 with overall 50% identical amino acids between the proteins. There is a distinct hydrophobic domain in the first 30 residues of the amino terminus of RhoGDIγ that is absent from the other two GDIs. Leucine made up 11 of the 30 residues.
Figure 2.
Comparison of amino acid sequence of RhoGDIγ with RhoGDI (RhoGDIα) and GDI/D4 (RhoGDIβ). Identical residues are indicated by straight lines, conserved residues are indicated by ∗. Conservative amino acids are grouped as follows: S, T, G, A, P; L, M, I, V; E, D, Q, N; R, K, H; F, Y, W; C. The hydrophobic domain in the amino-terminal region of RhoGDIγ is underlined.
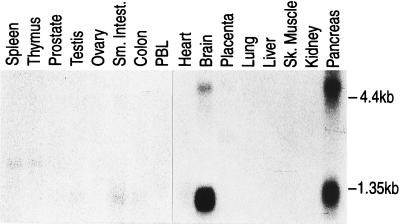
Northern blot analysis of poly(A)+ RNA from normal human tissues (Fig. 3) shows that the gene is expressed at high levels in the brain and pancreas. Two transcripts of 4.5 kb and 2.5 kb are detected with pancreatic tissues expressing about equal levels of both transcripts while brain tissue contains a higher level of the shorter transcript. Very low levels of the shorter mRNA is detected in small intestine and colon and a barely detectable level of the shorter mRNA is seen in ovarian and testicular tissues.
Figure 3.
Detection of RhoGDIγ mRNA in poly(A)+ RNA from normal human tissues. The filter (CLONTECH) contained 2 μg of poly(A)+ RNA per lane. The filter was hybridized with 32P-labeled RhoGDIγ cDNA probe, and washed with 0.2× standard saline citrate at 65°C before autoradiography for 24 hr at −80°C. Positions for 4.4 kb and 1.35 kb as marked by ribosomal markers are indicated.
Binding of RhoGDIγ to Rho-Related proteins.
Transfection of a GST–RhoGDIγ fusion cDNA into bacteria followed by induction revealed the production of a 52-kDa protein of the expected length. However the protein was present in inclusion bodies and was difficult to purify. As shown in Fig. 2, the amino terminus of the protein contained a hydrophobic domain that would explain the trapping of the protein in inclusion bodies. We therefore constructed a second fusion-protein vector with a deletion of the hydrophobic domain. Induction of bacteria transfected with the vector revealed ample expression of the protein and we were able to prepare sufficient quantity of the purified protein by glutathione agarose chromatography (3). This protein was used in subsequent functional studies.
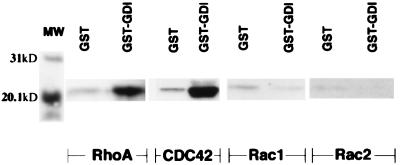
Insects cells (Sf9) infected with baculovirus vector containing CDC42, Rac1, Rac2, and RhoA were harvested and lysed. Cell lysates were then incubated with GST–RhoGDIγ in buffer (10 mM Tris, pH 7.4/150 mM NaCl in the presence of protease inhibitors) at room temperature for 3 hr. GST-fusion proteins were removed with glutathione agarose in batch preparation, rinsed and the beads separated on SDS/PAGE and Western blot analysis were prepared from the beads. Filters were then probed with antibodies (Santa Cruz Biotechnology) against RhoA, CDC42, Rac1, or Rac2. No binding was seen with Rac1 or Rac2 (Fig. 4). RhoGDIγ was able to bind to CDC42 and RhoA but with apparent lower affinity compared with RhoGDI (figures for RhoGDI binding to RhoA and related proteins not shown).
Figure 4.
Binding of RhoGDIγ to RhoA, CDC42, Rac1, and Rac2. Rho proteins were produced in insect cells infected with the various baculovirus as described in Materials and Methods. GST–RhoGDIγ bound to glutathione agarose beads was then incubated with lysates of insects cells for 18 hr at 4°C. GST alone was used as control. Bound proteins were separated by SDS/PAGE and Western filters were then probed with the respective antibodies (Santa Cruz Biotechnology) against RhoA, CDC42, Rac1, or Rac2. The figure shows binding of RhoA and CDC42 to RhoGDIγ (GST–GDI). The binding of Rac1 or Rac2 by GST–RhoGDIγ was the same as background binding by GST.
Assay for GDI Activity.
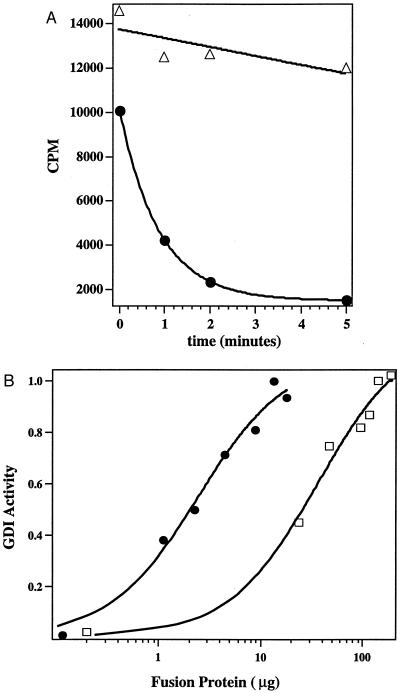
Using insect cells produced CDC42 as substrate, it can be seen that RhoGDIγ clearly inhibited the dissociation of GDP from the GTPase (Fig. 5A). To obtain a quantitative assessment of this activity, we derived the dose response curve for GDI activity against CDC42 comparing RhoGDIγ with RhoGDI. Fig. 5B shows the GDI activity of RhoGDIγ and RhoGDI against the CDC42 protein, determined in the same experiment. It can be seen that RhoGDIγ has 20–30 times less GDI activity against CDC42 than RhoGDI.
Figure 5.
Inhibition of GDP-dissociation from CDC42Hs by RhoGDIγ. (A) Time course of [3H]GDP dissociation from CDC42 upon mixing with low Mg2+ (7.5 mM EDTA) buffer in the presence of RhoGDIγ (▵) or control buffer (•). (B) Comparison of the relative efficiency of RhoGDI (•) and RhoGDIγ(□) in eliciting the GDI effect. CDC42Hs-[3H]GDP was incubated with the indicated amount of GDI, and the amount of radioactivity associated with CDC42Hs 5 min after initiating dissociation was determined by scintillation counter. The amount of the [3H]GDP still bound to the CDC42Hs protein was then normalized as a percentage of the amount bound in the presence of the maximum concentration of RhoGDI. The figure shows that the activity of RhoGDIγ is ≈20-fold less than that of RhoGDI.
Localization of RhoGDIγ by Immunofluorescence.
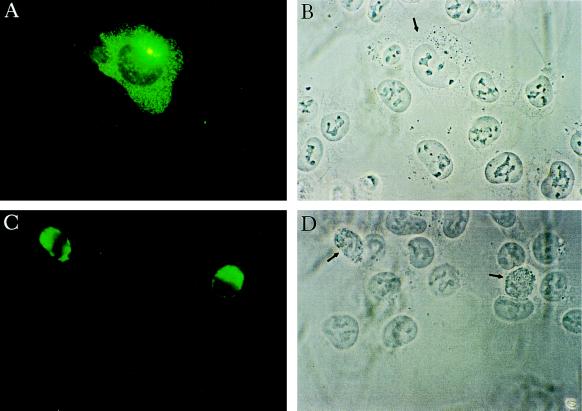
A GFP–RhoGDIγ fusion protein vector was constructed and transfected into baby hamster kidney cells. Twenty-four hours after transfection, two patterns of distribution were observed. In 60–80% of the cells, RhoGDIγ appeared as punctated proteins distributed homogeneously throughout the cytoplasm and with concentration in vesicles in the perinuclear area (Fig. 6A). In 20–40% of the transfected cells, the protein was distributed densely around the nuclei as dense irregular aggregates. These cells appeared spindly and some were rounded up. We then studied protein distribution in relation to cell morphology at timed intervals after transfection. At 8 hr transfected cells expressed a low level of the protein with a diffuse pattern. A few cells expressed high levels of protein and all these cells were rounded up. At 24–28 hr, a higher level of protein expression was seen in the cells, many with appearance as shown in Fig. 6A. However, there was also a higher percentage of rounded cells with very high levels of the protein as in Fig. 6C. By 60 hr, almost all the cells were rounded up and expressing high levels of RhoGDIγ. Furthermore, there was clearly a reduction in the percentage of positive cells. Staining with tetramethylrhodamine B isothiocyanate-labeled phalloidin showed that rounded cells have lost all stress fibers (figure not shown). Finally, costaining with Hoechst 33342 (Sigma) for nuclear DNA showed that in some of the cells there was hyperconvolution and fragmentation of the nuclei.
Figure 6.
Immunolocalization of RhoGDIγ. Plasmid DNA of GFP-tagged RhoGDIγ vector was transfected into baby hamster kidney cells grown on coverslips using the calcium phosphate precipitate method. At different times after transfection, the cells were fixed and the coverslips mounted in glycerol. Cells were examined under Argon arc light activation. (A) A cell expressing RhoGDIγ 24 hr after transfection, showing the typical diffuse punctate distribution of the protein with concentration in the perinuclear region. The yellowish spots are due to unavoidable overexposure from intensely fluorescent locations. (B) Same field as A taken under phase contrast. The positive cell is indicated by an arrow. (C) Two cells expressing very high levels of RhoGDIγ. (D) Same field as C under phase contrast, showing that the positive cells (arrows) are rounded up compared with neighboring normal adherent baby hamster kidney cells.
DISCUSSION
The tissue distribution of RhoGDIγ is quite different compared with that of RhoGDI or GDI/D4. Another difference is the presence of a long and short transcript of RhoGDIγ in the two tissues that express the highest levels of the mRNA (brain and pancreas). The full-length cDNA that we isolated contains an ORF that corresponds in length to that of the shorter transcript. The identity of the longer transcript is not known and could represent an alternatively spliced transcript. Another less likely possibility is that it represents the transcript of a highly homologous gene. Further work will be required to resolve this.
Alignment of the three GDIs shows that RhoGDIγ contains a distinct hydrophobic domain of ≈30 residues in the amino terminus (Fig. 2). This domain accounted for the considerable difficulty we encountered in our attempt to purify the full-length protein expressed in bacteria. Consequently we had to resort to the use of the truncated form with the hydrophobic domain deleted. Using this, we first demonstrated by an in vitro binding assay that RhoGDIγ differs from RhoGDI and GDI/D4 in that it is not able to bind to Rac1 or Rac2. To further substantiate this, we used the two-hybrid yeast system (18) to test for proteins that bind to RhoGDIγ. We find RhoGDIγ binds to CDC42 but not to Rac1 or Ras (data not shown).
Using an established in vitro assay for GDI activity, we next demonstrated that RhoGDIγ clearly functioned as a GDI against CDC42 (Fig. 5A) but with 20–30 times less efficacy compared with RhoGDI (Fig. 5B). This difference is very similar to the difference in GDI activity between RhoGDI and GDI/D47. In another previous study (15), we have shown that the deletion of as much as the first 20 residues of RhoGDI did not affect its GDI activity, whereas a deletion of as little as the last eight residues resulted in a total abrogation of the GDI activity of RhoGDI. Furthermore, a replacement of only a few amino acids in RhoGDI with the corresponding residues in GDI/D4 converted RhoGDI activity to that of GDI/D4 and vice versa. Thus the carboxyl end of the GDIs for Rho proteins appear to be a critical domain. In this region of RhoGDIγ, the residues (YLVVS) differ from that of either RhoGDI(YSIKS) or GDI/D4(YHNKS). Furthermore, the last eight residues of RhoGDIγ differ from RhoGDI and GDI/D4. Therefore it is not surprising that RhoGDIγ demonstrates a very different binding affinity for Rho and related proteins compared with RhoGDI. It is important to note that the GTPase-binding and GDI assays were done with the amino-terminal truncated form of RhoGDIγ because of the difficulty of purifying the whole protein from bacteria. Since the truncation did not affect the carboxyl end that is critical for binding to GTPase substrates, we conclude that the result we obtained with the amino-terminal truncated form of RhoGDIγ in the GDI assay very likely reflects the true level of GDI activity toward CDC42. Therefore, we conclude that RhoGDIγ is a GDI with a restricted spectrum of GDI activity compared with RhoGDI or GDI/D4. Whether RhoGDIγ will also bind to or function as a GDI for other related proteins such as RhoB, RhoC, and RhoG remains to be determined.
Recently it was shown that GDI/D4 is a substrate of the apoptosis protease (19). In hematopoietic cells induced to undergo apoptosis, GDI/D4 is truncated to a 23-kDa fragment at the CPP32 consensus cleavage site DELD (19)S. This consensus cleavage sequence is absent in both RhoGDI and RhoGDIγ.
RhoGDIγ tagged with GFP protein at the amino terminus showed a diffuse cytoplasmic distribution and in many cells the protein also was found clustered around the nucleus. A similar construction with GFP tagged to the amino terminus of GDI/D4 or RhoGDI showed only a diffuse distribution in the cytoplasm without any of the perinuclear aggregates seen with RhoGDIγ (figure not shown). The perinuclear pattern appears to resemble the staining pattern of organelles such as Golgi network and lysosomes but does not resemble endoplasmic reticulum. It is possible that the protein may become localized in certain intracellular compartments with the hydrophobic amino terminus of RhoGDIγ providing a mechanism for its membranes localization. Further colocalization studies with antibodies against proteins in various cellular compartments may reveal the nature of the location of RhoGDIγ. In this regard, it is of interest that RhoB was found to have a perinuclear distribution in contrast to RhoA and RhoC that are diffusely distributed in the cytoplasm and cell surface membrane (20). We are currently generating RhoB to study interactions with RhoGDIγ.
Members of the Rho-related proteins appear to be critical in cytoskeletal organization such as the formation of stress fibers, membrane ruffling (21, 22) and the formation of filopodia (23).
A high level of expression of RhoGDIγ caused cells to round up. The reduction in the percentage of positive cells at 60 hr after transfection of the expression vector may be explained by the loss of rounded cells which had become detached. Consistent with our observation that the RhoGDIγ binds to RhoA, an over expression of the protein would lead to an inhibition of activation of RhoA and hence the loss of stress fibers. The phenotype is similar to that seen with the injection and overexpression of RhoGDI on cell morphology (10) or inhibitors of RhoA such as the exoenzyme C3 transferase from Clostridium botulinum (21, 22). In some of the rounded cells, the hyperconvolution of nuclei and DNA fragmentation observed is consistent with apoptosis but more elaborate studies will be required to determine whether or not apoptosis can be induced by the overexpression of RhoGDIγ.
The significance of the preferential expression of RhoGDIγ gene in brain and pancreas awaits future studies. A cellular function that both tissues are actively engaged in is the packaging and secretion of proteins in vesicles; neurotransmitters in synaptic junctions and hormones in exocrine or endocrine cells. Thus, RhoGDIγ may be involved in secretory pathways.
The data presented here showed that RhoGDIγ is another member of an emerging family of regulators for the Rho-related proteins. It is possible that, similar to the GDP exchange factors for the Rho proteins, there may be a large family of these GDIs. Within the family of GDP exchange factors for the Rho proteins, some are specific exchange factors for individual Rho proteins [e.g., Lbc (24), TIAM-1 (25)] while others are GDP exchange factors for more than one of the proteins [e.g., Dbl (26), Vav]. Thus far, the GDIs for the Rho proteins have shown no substrate specificity by in vitro assays. Furthermore the same cell may contain more than one GDIs. Whether and how they each function to exert distinct cellular effects will require more elaborate in vivo studies. For example, even though in vitro assays indicate that RhoGDI is a GDI for the Rho, Rac, and CDC42 proteins, the injection of RhoGDI into cells affect dramatically the formation of stress fibers, a function mediated by RhoA. Injected cells still retain ability to form lamellipodia, a function mediated by Rac proteins (data not shown). This would suggest that there is a preferential in vivo effect of RhoGDI on RhoA rather than Rac proteins. In contrast, the subtle defect in superoxide production in macrophages with loss of function of GDI/D4 points to an important role of GDID4 in regulation of Rac proteins (16). The normal phenotype of animals with loss of function of GDI/D4 also suggest that there may be considerable redundancy of function between the RhoGDIs. The identification of RhoGDIγ should widen the scope of experiments and database to understand the physiological importance of these GDIs. Finally, to avoid confusion and to facilitate communication, we propose that RhoGDI be renamed RhoGDIα, and GDI/D4 be named RhoGDIβ.
Acknowledgments
We thank Dr. Christopher Carpenter (Harvard Medical School) for the RhoA baculovirus and Dr. Gary Bokoch (Scripps Research Institute) for the Rac1 and Rac2 baculovirus. This work is supported by Grant DHP-4B (B.L. and C.N.A.) from the American Cancer Society and Grant RO1 DK47636 (B.L. and C.N.A.) from the National Institutes of Health. B.L. is a Scholar of the Leukemia Society of America.
ABBREVIATIONS
- GDI
GDP-dissociation inhibitor
- GST
glutathione S-transferase, GFP, green fluorescent protein
Note
After this manuscript was submitted for review, a homologous murine protein named RhoGDI3 was described (27). The protein is >80% identical to RhoGDIγ and is very likely the murine equivalent of RhoGDIγ. RhoGDI3 was found to bind to RhoB and RhoG. RhoGDI3 was also shown to function as a GDI for RhoB. However, contrary to what we observed, RhoGDI3 did not appear to bind to RhoA. Furthermore the interaction of RhoGDI3 with CDC42 was not examined. The combined data indicate that RhoGDIγ may bind with and function as a GDI for CDC42, RhoB, and RhoG and possibly RhoA.
Footnotes
References
- 1.Boguski M S, McCormick F. Nature (London) 1993;366:643–654. doi: 10.1038/366643a0. [DOI] [PubMed] [Google Scholar]
- 2.Fukumoto Y, Kaibuchi K, Hori Y, Fujiko H, Araki S, Ueda T, Kikuchi A, Takai Y. Oncogene. 1990;5:1321–1328. [PubMed] [Google Scholar]
- 3.Leonard D, Hart M, Platko J, Eva A, Henzel W, Evans T, Cerione R A. J Biol Chem. 1992;267:22860–22868. [PubMed] [Google Scholar]
- 4.Abo A, Pick E, Hall A, Totty N, Teahan C, Segal A. Nature (London) 1991;353:668–670. doi: 10.1038/353668a0. [DOI] [PubMed] [Google Scholar]
- 5.Knaus U, Keyworth P, Kinsella B, Curnette J, Bokoch G M. J Biol Chem. 1992;267:23575–23582. [PubMed] [Google Scholar]
- 6.Lelias J M, Adra C N, Wulf G M, Guillemot J C, Khagad K, Caput D, Lim B. Proc Natl Acad Sci USA. 1993;90:1479–1483. doi: 10.1073/pnas.90.4.1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Adra C N, Ko J, Leonard D, Wirth L J, Cerione R A, Lim B. Genes Chromosomes Cancer. 1993;8:253–261. doi: 10.1002/gcc.2870080408. [DOI] [PubMed] [Google Scholar]
- 8.Scherle P, Behrens T, Staudt L M. Proc Natl Acad Sci USA. 1993;90:7568–7572. doi: 10.1073/pnas.90.16.7568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Takaishi K, Kikuchi A, Kuroda S, Kotani K, Sasaki T, Takai Y. Mol Cell Biol. 1993;13:72–79. doi: 10.1128/mcb.13.1.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Miura Y, Kikuchi A, Musha T, Kuroda S, Yaku H, Sasaki T, Takai Y. J Biol Chem. 1993;268:510–515. [PubMed] [Google Scholar]
- 11.Leffers H, Nielsen M S, Andersen A H, Honore B, Madsen P, Vandekerckhove J, Celis J E. Exp Cell Res. 1993;209:165–174. doi: 10.1006/excr.1993.1298. [DOI] [PubMed] [Google Scholar]
- 12.Hart M J, Maru Y, Leonard D, Witte O N, Evans T, Cerione R A. Science. 1992;258:812–815. doi: 10.1126/science.1439791. [DOI] [PubMed] [Google Scholar]
- 13.Chuang T H, Xu X, Knaus U G, Hart M J, Bokoch G M. J Biol Chem. 1993;268:775–778. [PubMed] [Google Scholar]
- 14.Hancock J, Hall A. EMBO J. 1993;12:1915–1921. doi: 10.1002/j.1460-2075.1993.tb05840.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Platko J, Leonard D, Adra C N, Shaw R, Cerione R, Lim B. Proc Natl Acad Sci USA. 1995;92:2974–2978. doi: 10.1073/pnas.92.7.2974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guillemot J-C, Kruskal B A, Adra C N, Zhu S, Ko J L, Burch P, Nocka K, Seeto K, Simons E, Lim B. Blood. 1997;88:2722–2731. [PubMed] [Google Scholar]
- 17.Sambrook J, Fritsch E, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 18.Fields S, Song O. Nature (London) 1998;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- 19.Na S, Chuang T-H, Cunningham A, Tui T G, Hankel J H, Bokoch G M, Danley D E. J Biol Chem. 1996;271:11209–11213. doi: 10.1074/jbc.271.19.11209. [DOI] [PubMed] [Google Scholar]
- 20.Zalcman G, Closson V, Linares-Cruz G, Lerebours F, Honore N, Tavitian A, Olofsson B. Oncogene. 1995;10:1935–1945. [PubMed] [Google Scholar]
- 21.Ridley A J, Hall A. Cell. 1992;70:389–399. doi: 10.1016/0092-8674(92)90163-7. [DOI] [PubMed] [Google Scholar]
- 22.Ridley A J, Paterson H F, Johnston C L, Diekmann D, Hall A. Cell. 1992;70:401–410. doi: 10.1016/0092-8674(92)90164-8. [DOI] [PubMed] [Google Scholar]
- 23.Nobes C D, Hall A. Cell. 1995;81:53–62. doi: 10.1016/0092-8674(95)90370-4. [DOI] [PubMed] [Google Scholar]
- 24.Zheng Y, Olson M F, Hall A, Cerione R A, Toksoz D. J Biol Chem. 1995;270:9031–9034. doi: 10.1074/jbc.270.16.9031. [DOI] [PubMed] [Google Scholar]
- 25.Michiels F, Habets G G, Stam J C, van der Kammen R A, Collard J G. Nature (London) 1995;375:338–340. doi: 10.1038/375338a0. [DOI] [PubMed] [Google Scholar]
- 26.Yaku H, Sasaki T, Takai Y. Biochem Biophys Res Commun. 1994;198:811–817. doi: 10.1006/bbrc.1994.1116. [DOI] [PubMed] [Google Scholar]
- 27.Zalcman G, Closson V, Camonis J, Honore N, Rousseau-Merck M-F, Tavitian A, Olofsson B. J Biol Chem. 1996;271:30366–30374. doi: 10.1074/jbc.271.48.30366. [DOI] [PubMed] [Google Scholar]