Abstract
In eukaryotic cells, accumulation of unfolded protein in the endoplasmic reticulum induces transcription of a family of genes encoding endoplasmic reticulum protein chaperones through a conserved unfolded protein response element. In Saccharomyces cerevisiae, activation of a transmembrane receptor kinase, Ire1p (Ern1p), initiates signaling, although the mediators immediately downstream of Ire1 kinase are unknown. Here we demonstrate interaction of Ire1p with the transcriptional coactivator, Gcn5p (for general control nonrepressed; also known as Ada4p). Gcn5p associates with other Ada (for alteration/deficiency in activation) gene products in a heteromeric complex and has histone acetyltransferase activity. We show that the Gcn5/Ada complex is selectively required for the unfolded protein response but not for the heat shock response. A novel mechanism is proposed in which activation of a receptor kinase recruits a transcription coactivator complex to a specific chromosomal locus to mediate localized histone acetylation, thus making specific gene sequences accessible for transcription.
Keywords: stress response, Gcn5, Ada complex, IRE1, KAR2
The endoplasmic reticulum (ER) in all eukaryotic cells is the site where protein folding and assembly occurs for secreted and transmembrane proteins. Protein folding in the ER is assisted by protein chaperones that either prevent nonproductive reactions to limit protein aggregation or assist protein folding (1). The expression of these protein chaperones is regulated according to their needs. For example, accumulation of unfolded proteins or mutant proteins in the ER initiates a signal to activate transcription of genes such as BiP, protein disulfide isomerase, and the 94-kDa glucose-regulated protein GRP94. A conserved promoter element, unfolded protein response element (UPRE), was identified in yeast (2) and mammalian cells (3) that is necessary and sufficient to mediate the transcriptional induction in response to unfolded protein in the ER. In the yeast Saccharomyces cerevisiae, transcription of KAR2, the yeast homologue for mammalian BiP, is also controlled (2) by a heat shock response element (HSE) and a G+C-rich region that contributes to high level of constitutive expression. Recently, it was shown that the binding of a basic leucine zipper protein, Hac1p, at the UPRE is required for the transcriptional induction of KAR2 in response to unfolded proteins in the ER (4).
The transcriptional induction of KAR2 by unfolded proteins also requires a transmembrane Ser/Thr kinase, Ire1p (5, 6). Ire1p is structurally similar to class I growth factor receptors and has three distinct domains, an N-terminal ER luminal domain, a transmembrane domain that spans the ER membrane, and a C-terminal kinase domain that is either in the cytoplasm or in the nucleoplasm. The cytoplasmic/nucleoplasmic domain has intrinsic Ser/Thr kinase activity (7), and Ire1p undergoes autophosphorylation in response to unfolded proteins in the ER (8). Thus, Ire1p appears to be the proximal sensor of unfolded proteins in the ER that initiates the unfolded protein response (UPR).
In eukaryotes, transcriptional activation requires functional interactions between activators bound to the upstream activating sequences and the general transcription factors that occupy the TATA box. It has been proposed that these interactions are mediated through a set of factors termed coactivators or adaptors. In the yeast S. cerevisiae, one set of coactivators, including Gcn5p, Ada2p, Ada3p, and Ada5p (Spt20p), is known to exist in a multimeric complex (Gcn5/Ada) and is required for the maximal activation of a subset of acidic activators (9–12). In spite of numerous interaction studies and genetic data, the biochemical mechanism of action of these components is largely unknown. Recently, Gcn5p was shown to have histone acetyltransferase activity (13), providing one biochemical function for the complex and an explanation for the long-existing relationship between histone acetylation and gene activation.
To understand how the Ire1 kinase transmits the unfolded protein signal, we searched for the downstream signaling molecules. In this paper, we demonstrate that the cytoplasmic domain of Ire1 kinase interacts with Gcn5p both in vivo and in vitro and that the Gcn5/Ada complex is selectively required for the UPR but not for the heat shock response (HSR). This observation provides a link between the proximal sensor for the UPR and the mechanism of selective transcriptional activation. We propose a novel mechanism in which activation of a transmembrane kinase recruits a transcription coactivator complex to a specific chromosomal locus to mediate localized histone acetylation, thus making specific gene sequences accessible for transcription.
MATERIALS AND METHODS
Yeast Strains, General Methods, and Plasmid Constructions.
The Escherichia coli strain DH5α was used for the propagation of plasmids. The genotypes of S. cerevisiae strains used in this study were as follows: EGY48, Matα his3 leu2::3LexAop-LEU2 ura3 trp1 LYS2 (14); and BWG1-7a, Matα leu2-3,112 his4-519 ade 1-100 ura3-52 (11). The genetic methods and standard media were previously described (15). The ire1 deletion was generated by PCR-based gene disruption method described by Wach et al. (16). The BWG1-7a strain was transformed with PCR-generated DNA molecules carrying the kanr gene flanked by 50 bases of IRE1 homology using 5′ primer 5′-AACTTCAGGAATGTGAAAATATGATTGTAATAGGCAAAACTATTTTTGAGGGCCACTAGTGGATCTGA-3′ and 3′ primer 5′-AACGGATCGTGATCAATCATTTGGGAGATCAGATCTGTAGCTTCTGCAATAAGCTTCGTACGCTGCAG-3′. G418-resistant colonies were isolated and Δire1 cells that lack a 670-aa region of Ire1p, including the transmembrane and the kinase domain, were confirmed by Southern blotting. To construct UPRE–LacZ reporter, the 4-kb BglII–TthIII fragment of pJC005 (5) containing the UPRE, crippled CYC1 promoter, and lacZ coding sequence was subcloned into the BamHI and NaeI sites of pRS315 (17). Construction of the fusions between Ire1 cytoplasmic domain and LexA DNA binding domain (LexADB) was previously described (7). pEG22-A carrying the LexA–Cdc28p fusion was generously provided by R. Brent (18). The bromodomain (BD) was amplified by PCR from pDB20LHA-GCN5 (10) using 5′ primer 5′-CTACTCGAGGCTTGGCCCTTCTTACAACCC-3′ and 3′ primer 5′-TTCCTCGAGTTAATCAATAAGGTGAGAATATTCAGG-3′ and subcloned into the XhoI site of pJG4-5 (14). The yeast two-hybrid assays (14) and yeast transformations (19) were performed as described previously.
Immunoprecipitations, in Vitro Binding Assays, and Western Blot Analysis.
Yeast cell lysates were made according to Williams et al. (20). Construction, expression, and purification of the glutathione S-transferase (GST)–Ire1p fusion protein and the in vitro binding assay were previously described (7). Western blotting was performed by standard procedures (21) using anti-influenza hemagglutin epitope (HA) primary antibodies (Boehringer Mannheim) and horseradish peroxidase-conjugated goat anti-mouse secondary antibodies (GIBCO/BRL). Bands were detected using the enhanced chemiluminescence (ECL) kit (Amersham) and quantified using the National Institutes of Health program image. Yeast cell lysates containing equal amounts of different B42–HA–Gcn5 fusion proteins were immunoprecipitated in RIPA buffer (21).
Northern Blot Analysis.
Isolation of total RNA (22) and Northern blot hybridization analysis (23) were performed according to standard protocols. Blots were sequentially probed with a random primer-labeled 1.2-kb HindIII fragment of KAR2, a 0.6-kb fragment of PDI1, and a 1-kb HindIII fragment of the ACT1 gene, which encodes actin in S. cerevisiae. KAR2, PDI1, and ACT1 mRNAs were quantified by PhosphoImager scanning (Molecular Dynamics). RNA from tunicamycin-treated cells was normalized to ACT1 mRNA. Because the ACT1 mRNA is heat-labile, RNA from the heat-shocked cells was normalized to rRNA that was quantified by scanning the ethidium bromide-stained gels using image.
RESULTS
Gcn5p Interacts with Ire1p in Vivo and in Vitro.
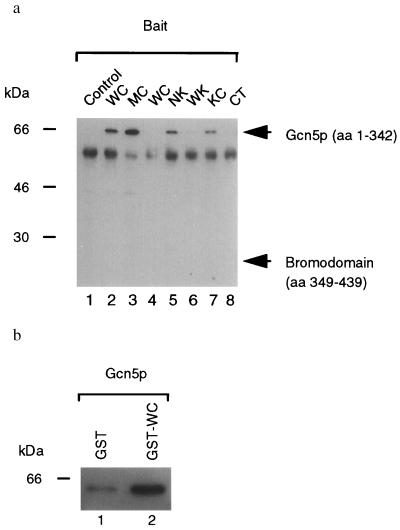
To identify mediators of downstream signaling from Ire1p, we used a modified version of the yeast two-hybrid system (14). The cytoplasmic/nucleoplasmic domain of Ire1p containing the intact kinase domain was used to screen a yeast genomic library. The kinase that shows the highest homology to Ire1p, Cdc28p, was used as a negative control to eliminate nonspecific interactions with Ser/Thr kinases. GCN5 was isolated in this screen as a specific interactor of Ire1p that did not interact with Cdc28p (Fig. 1a). The genetic interaction in the two-hybrid system was confirmed by detecting a physical interaction between the LexA–Ire1p and B42–HA–Gcn5p fusion proteins by coimmunoprecipitation of yeast extracts using anti-LexA antibody and Western immunoblot analysis of the immunoprecipitates using anti-HA antibody (Fig. 2a, lanes 1 and 2). E. coli-expressed GST–Ire1p fusion protein was used in an in vitro binding assay to immunoabsorb B42–HA–Gcn5p from yeast cell lysates. Subsequently, the absorbed proteins were detected by Western analysis using the anti-HA antibody. This assay revealed that B42–HA–Gcn5p associated to a greater degree with GST–Ire1p (9-fold) compared with the control, GST, alone (Fig. 2b), demonstrating a physical interaction between Gcn5p and Ire1p. In the coimmunoprecipitation experiments (Fig. 2a, lane 3), two-hybrid assays (data not shown), and in vitro binding assays (data not shown), the kinase-defective K702A mutant cytoplasmic/nucleoplasmic domain also interacted with Gcn5p. Although these studies were not designed to measure binding affinities of Gcn5p with the wild-type or the mutant kinase, they suggested that Gcn5p can interact with both phosphorylated and dephosphorylated forms of the receptor. Alternatively, Gcn5p interacted only with the phosphorylated form of Ire1p, and the mutant cytoplasmic/nucleoplasmic domain bait was phosphorylated in vivo by the endogenous Ire1p.
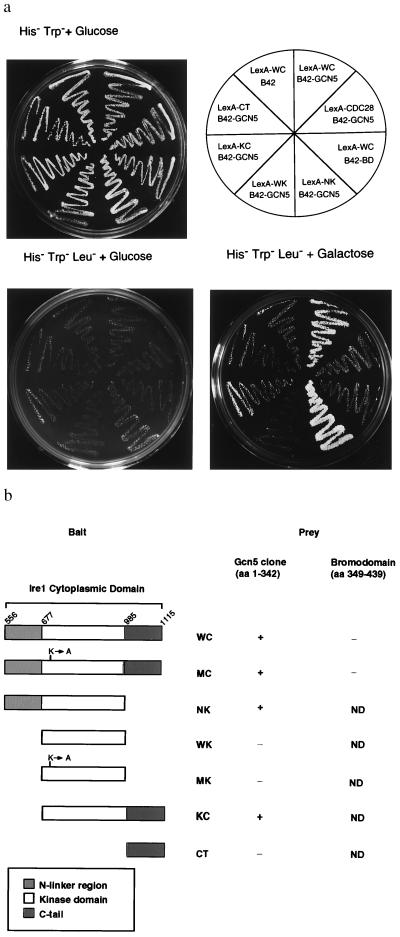
Figure 1.
Analysis of Ire1p interaction with Gcn5p and mapping of interaction domains. (a) LexA fusion proteins of the cytoplasmic domain and different subdomains within the cytoplasmic domain of Ire1p (bait) were tested for interaction with either the original clone B42–HA–Gcn5p (amino acids 1–342) or with B42–HA–Gcn5 BD (amino acids 349–439). Transformants harboring IRE1 and GCN5 fusions were patched onto His−Trp− and replica-plated onto His−Trp−Leu− plates containing either glucose or galactose. (b) Diagrammatic representation of the IRE1 deletions and their in vivo genetic and physical interaction activities. One-letter abbreviations for amino acids are used. WC, wild-type cytoplasmic/nucleoplasmic domain; MC, K702A mutant cytoplasmic/nucleoplasmic domain; NK, N-linker plus kinase domain; WK, wild-type kinase domain; MK, K702A mutant kinase domain; KC, kinase plus C-terminal tail; CT, C-terminal tail; ND, not detected.
Figure 2.
Physical association of Ire1p and Gcn5p in vivo and in vitro. (a) Coimmunoprecipitations of Gcn5p from yeast cell lysates. Lysates from cells coexpressing either B42–HA–Gcn5 (amino acids 1–342; lanes 1–3 and 5–8) or B42-HA-Gcn5 BD (amino acids 349–439; lane 4) as prey and LexADB (lane 1), LexADB–WC (lanes 2 and 4), LexADB–MC (lane 3), LexADB–NK (lane 5), LexADB–WK (lane 6), LexADB–KC (lane 7), and LexADB–CT (lane 8) as bait were immunoprecipitated with anti-LexA antibodies (generously provided by Erica Golemis, Fox Chase Cancer Center, Philadelphia), and immunoprecipitated proteins were analyzed by Western blotting with anti-HA antibody (Boehringer Mannheim). Expected migration of the prey proteins are indicated. The 65-kDa species is the size expected for the B42 transcriptional activator–HA–Gcn5p (residues 1–342) fusion protein. The 55-kDa background band is rabbit IgG heavy chain. WC, wild-type cytoplasmic/nucleoplasmic domain; MC, K702A mutant cytoplasmic/nucleoplasmic domain; NK, N-linker plus kinase domain; WK, wild-type kinase domain; KC, kinase plus C-terminal tail; CT, C-terminal tail. (b) In vitro binding assay. Either GST (lane 1) or GST–WC (lane 2) proteins were used to immunoabsorb B42–HA–Gcn5p and absorbed proteins analyzed by Western blotting with anti-HA antibody (Boehringer Mannheim). WC, wild-type cytoplasmic/nucleoplasmic domain.
Gcn5p contains an acetyltransferase catalytic core at the N terminus (13) and a BD at the C terminus (amino acids 349–422; ref. 9) that is thought to mediate protein–protein interactions and to tether Gcn5p to other factors bound to specific chromosomal sites. However, the BD is not required for Gcn5p–Ire1p interaction, as the GCN5 clone isolated by the two-hybrid analysis did not contain the BD. Moreover, the BD alone did not interact with Ire1p in the two-hybrid assay (Fig. 1a) nor did it show physical association with Ire1p (Fig. 2a, lane 4). These results indicate that the N-terminal portion of Gcn5p that contains the acetyltransferase core interacts with Ire1p. To dissect the molecular details of the Ire1p–Gcn5p interaction, C-terminal and/or N-terminal truncations were made in the Ire1p cytoplasmic/nucleoplasmic domain (see Fig. 1b). Although these assays are not designed to measure binding affinities, deletion of the N-linker region (amino acids 556–676; kinase plus C-terminal tail) apparently reduced, but did not destroy, either the genetic (Fig. 1a) or the physical interaction (Fig. 2a, lanes 5 and 7). In contrast, deletion of the C-terminal tail (amino acids 986-1115; N-linker plus kinase domain) did not reduce the interaction. Moreover, expression of either the kinase domain alone or the C-terminal tail alone was not sufficient to mediate interaction with Gcn5p (Fig. 1a; Fig. 2a, lanes 6 and 8). Although we cannot rule out that the isolated kinase domain (wild-type kinase domain) had an altered secondary structure that did not permit interaction, these results suggest that there is more than one Gcn5p interaction site within Ire1p; perhaps such sites are scattered throughout cytoplasmic/nucleoplasmic domain, and multiple sites are required for the detectable interaction.
Recently, we and others (7, 8) have shown that the cytoplasmic/nucleoplasmic domain of Ire1p has intrinsic Ser/Thr kinase activity and the activation of the Ire1p function requires oligomerization and trans-phosphorylation of the cytoplasmic/nucleoplasmic domain. To test whether Gcn5p is a substrate for Ire1p kinase, we exhaustively looked for phosphorylation of Gcn5p in vitro using bacterially expressed Gcn5p and GST–Ire1p. However, Gcn5p phosphorylation by Ire1p was not detected.
Gcn5p Is Required for the UPR.
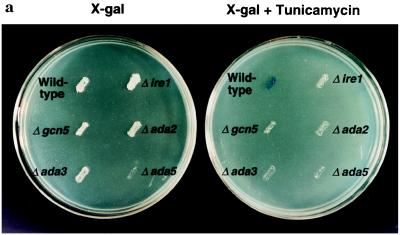
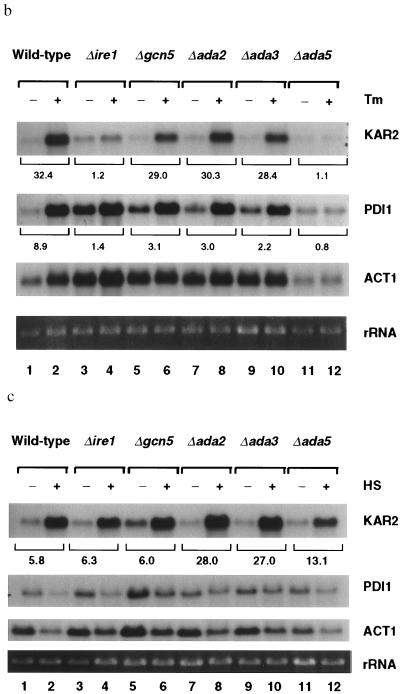
If the interaction between Ire1p and Gcn5p was physiologically significant for transcriptional induction mediated through the UPRE, we would expect that cells deficient in Gcn5p would be defective in the UPR. To test this hypothesis, we studied the UPR in a Δgcn5 strain harboring the 22-bp UPRE element upstream from a lacZ reporter gene. The UPR was monitored on 5-bromo-4-chloro-3-indolyl β-d-galactoside (X-Gal) plates either with or without tunicamycin, a drug that inhibits N-linked glycosylation, thus leading to accumulation of unfolded proteins in the ER. Wild-type cells turned blue on X-Gal plates containing tunicamycin within 24 hr, indicating an intact UPR pathway (Fig. 3a). In contrast, the isogenic Δgcn5 cells remained white, similar to Δire1 cells. These results show that GCN5 is required for transcriptional induction in response to unfolded proteins mediated through the UPRE (Fig. 3a). However, upon longer incubations (48 hr) on X-Gal plates, Δgcn5 cells turned light blue compared with wild-type cells, which were dark blue, indicating that the UPR is partially defective in Δgcn5 cells. To characterize the effect of Gcn5p on the intact endogenous KAR2 promoter that contains multiple promoter elements, we evaluated the induction of KAR2 (BiP) mRNA in wild-type and Δgcn5 cells in response to unfolded proteins in the ER by Northern blot analysis and PhosphoImager quantitation. Upon tunicamycin treatment, in comparison to wild-type cells (Fig. 3b, lanes 1 and 2), transcriptional induction of KAR2 was reproducibly reduced by ≈10% in Δgcn5 cells (Fig. 3b, lanes 5 and 6) in three independent experiments. To investigate whether the requirement of Gcn5p for the UPR is a phenomenon limited to KAR2, we measured the mRNA levels of PDI1, a gene that encodes another ER chaperon protein (24) and is transcriptionally induced by tunicamycin (5). Interestingly, the transcriptional induction of PDI1 was not only defective in Δgcn5 cells (3.1-fold) compared with the wild-type cells (8.9-fold), but also it was more pronounced compared with the transcriptional induction of KAR2. This apparent difference in the requirement for Gcn5p in the induction of KAR2 and PDI1 could be due to a higher level of basal expression of PDI1 and/or a lower level of PDI1 induction compared with KAR2. These results indicate that Gcn5p is required for the maximal activation of the UPR.
Figure 3.
The Gcn5/Ada coactivator complex is selectively required for the unfolded protein response. (a) Monitoring the UPR. Isogenic yeast strains in BWG1-7a background were transformed with UPRE–lacZ reporter construct. Transformants were replica-plated onto Leu− medium containing X-Gal, either with or without tunicamycin (2 μg/ml). Plates were incubated for 24 hr at 30°C. (b) Nothern blot analysis of total yeast RNA from tunicamycin-treated cells. Yeast cultures grown in liquid yeast extract/peptone/dextrose (YPD) medium to early logarithmic phase were further incubated for 90 min at 30°C with (+) or without (−) tunicamycin (2 μg/ml), and the transcriptional induction of KAR2 and PDI1 was assayed by Northern blot analysis. KAR2 and PDI1 mRNA abundances were quantified by PhosphoImager scanning and normalized to actin mRNA, and the fold inductions are indicated. Tm, tunicamycin. (c) Northern blot analysis of total yeast RNA from heat-shocked cells. Yeast cultures grown at 23°C in liquid YPD medium to early logarithmic phase were further grown for 15 min either with (+) or without (−) heat shocking at 39°C. The induction of KAR2 and PDI1 transcription was assayed by Northern blot analysis. The abundances of KAR2 and PDI1 mRNAs were determined by PhosphoImager scanning and normalized to rRNA, and the data are indicated as fold induction. HS, heat shock.
Ada2p, Ada3p, and Ada5p Are Also Required for the UPR.
As Gcn5p associates with Ada2p, Ada3p, and Ada5p in a heteromeric complex that is required for the maximal activation of a subset of acidic transcriptional activators (9, 10, 12), we measured the unfolded protein response using the same reporter assay in Δada2, Δada3, and Δada5 cells otherwise isogenic to wild-type strain, BWG1-7a. Induction of the UPR in these deletion strains, as well as a Δada2/Δada3 double deletion strain (data not shown), was also defective (Fig. 3a). Similar to the Δgcn5 cells, upon longer incubations (48 hr), the Δada2, Δada3, and Δada2/Δada3 cells turned light blue, whereas Δada5 cells and Δire1 cells remained white. In agreement with these results, Northern blot analysis (Fig. 3b lanes 7–10) repeatedly demonstrated about 10% reduced induction of KAR2 mRNA and a greater reduction in PDI1 mRNA in tunicamycin-treated Δada2 and Δada3 cells. The induction of both KAR2 and PDI1 was completely defective in Δada5 cells. KAR2 was induced by only 1.1-fold in Δada5 cells in response to unfolded protein (Fig. 3b, lanes 11 and 12), which was comparable to a 1.2-fold induction in Δire1 cells (Fig. 3b, lanes 3 and 4), but was in sharp contrast to the wild-type cells, which showed a 32.4-fold induction. These results demonstrate that the majority of the transcriptional induction in response to Ire1p activation is mediated through Ada5p and underscore the requirement for Ada5p in KAR2 transcription in response to unfolded protein in the ER. These results also show that the Gcn5/Ada complex is required globally for the UPR.
Gcn5/Ada Complex Is Not Required for the HSR.
Because KAR2 also contains a HSE that is functionally distinct from the UPRE (2), we asked whether the HSE also requires the Gcn5/Ada complex for activation of the KAR2 promoter in response to heat shock. Interestingly, the deletion of GCN5 had no effect on heat-mediated KAR2 induction, whereas the deletion of ADA2 and ADA3 brought about an increased KAR2 induction (Fig. 3c). Moreover, Δada5 cells that showed 29.4-fold reduction in the UPR retained inducibility of KAR2 in response to heat shock (13.1-fold; Fig. 3c, lanes 11 and 12) compared with wild-type cells (5.8-fold; Fig. 3c, lanes 1 and 2). These results demonstrate that Gcn5/Ada complex is selectively required for the transcriptional induction of KAR2 mediated through the UPRE but not through the HSE. In addition, these results also indicate that the inability of the deletion strains (Δada5 cells in particular) to maximally activate the UPR is not due to a generalized defect in transcriptional induction, because the HSR is intact in these strains.
DISCUSSION
We have isolated Gcn5p as a specific interactor of Ire1p kinase and have provided evidence for both genetic and physical association between the two proteins. To our knowledge, this is the first demonstration of a signaling molecule immediately downstream of the transmembrane Ire1p kinase and upstream of the transcriptional coactivator Gcn5p. Gcn5p is a component of the multisubunit complex Gcn5p/Ada2p/Ada3p/Ada5p (Gcn5/Ada; refs. 11 and 12) and was recently shown to exhibit histone acetyltransferase activity. Ada3p appears to hold the complex together by virtue of its interactions with Gcn5p, Ada2p, and Ada5p. Ada2p physically interacts with both acidic activation domains and TATA-binding proteins (25). Ada5p (Spt20p) also interacts with at least the acidic activator domain of VP16 and is required for normal TATA-binding function at certain promoters (26). We have also shown that the Gcn5/Ada complex is required for the transcriptional induction of KAR2 in response to accumulation of unfolded proteins in the lumen of the ER. These findings support a physiological significance for the Ire1p–Gcn5p interaction. The null mutants of both ire1 and ada5 are qualitatively and quantitatively similar in their UPR and have a common inositol-requiring phenotype (26, 27), suggesting that both IRE1 and ADA5 function in a common pathway. Moreover, the selective requirement for the Gcn5p complex to activate the UPR but not the HSR demonstrates a differential Gcn5/Ada5 complex dependency of the UPRE and HSE elements within the KAR2 promoter in responding to two different stresses. Unexpectedly, Δada strains exhibited an increased heat shock induction of KAR2 compared with wild-type strain. Induced expression of the heat shock genes is controlled at the level of transcript elongation (28, 29). Because Gcn5/Ada-dependent transcription is abrogated in cells lacking these coactivators, RNA polymerase and/or other factors become more available to transcription units that do not require the Gcn5/Ada complex. This may explain the increased KAR2 induction in response heat observed in Δada2, Δada3, and Δada5 cells.
Acetylation of histones is a landmark of transcriptionally active chromatin (30), whereas deacetylation of histones correlates with transcriptional silencing (31). The amount of histone acetylation is determined by an equilibrium between acetylase and deacetylase activities, suggesting that chromatin structure could be reversibly modified by targeting histone acetylase and deacetylase to a specific gene. On the basis of nuclear localization of Ada5p (26) and the enrichment of Gcn5p in the nucleus (13), it appears that the Gcn5/Ada complex is nuclear. Although the N-terminal portion of the Ire1p has been localized to the lumen of the ER (6), localization of the C terminus following the transmembrane domain is not known. The C terminus of Ire1p may exist in the nucleoplasm, because the ER membrane is a continuation of the nuclear envelope. Data presented here suggest that the molecular alterations due to Ire1p–Gcn5p interactions leads to targeting of the Gcn5/Ada complex to UPRE-containing genes encoding the ER chaperons, including KAR2 and PDI1 (Fig. 4). In contrast, activation of the KAR2 promoter through the HSE occurs through a different pathway and does not require the Gcn5/Ada complex.
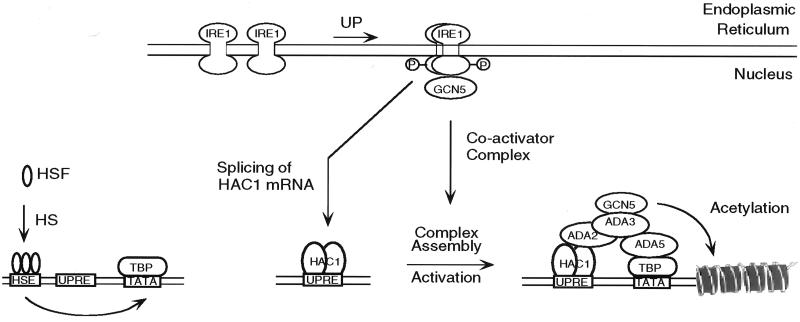
Figure 4.
Gcn5/Ada complex-dependent and -independent pathways of selective transcriptional activation. UPR and HSR activations of the KAR2 promoter are depicted as independent signaling pathways in S. cerevisiae. UP, unfolded proteins; HS, heat shock; HSF, heat shock factor; TBP, TATA-binding protein.
In S. cerevisiae, the UPRE is occupied by an ER stress-inducible transcription factor, Hac1p. Approximately two-thirds of the Ire1p-dependent transcriptional activation of UPRE-containing promoters is mediated through Hac1p (4). Hac1p is a basic leucine zipper transcriptional activator similar to Gcn4p, a transcriptional activator for amino acid biosynthetic genes that are induced under the stress condition of amino acid starvation. The Gcn5/Ada complex may act through Hac1p analogous to its mechanism of activation with Gcn4p (9, 10) to permit maximal transcriptional activation. Experiments are presently in progress to determine whether the Gcn5/Ada-dependent activation of the UPR requires Hac1p. In contrast to Δgcn5, Δada2, and Δada3 cells, in which the UPR is only partially defective, Δada5 cells were completely defective for the UPR and similar Δire1 cells. However, both Δire1 and Δada5 cells were more defective in the UPR than Δhac1 cells (4). Thus, we conclude that the Hac1p-dependent transcriptional activation of the UPR requires Ada5p (Fig. 4).
To date, we have not detected phosphorylation of Gcn5p by Ire1p kinase in vitro. If phosphorylation by Ire1p is important in regulating the activity of the Gcn5p/Ada complex, it is possible that Gcn5p is the Ire1p-interacting partner of the complex, and a different subunit is the substrate for phosphorylation. Alternatively, Gcn5p may be a substrate for Ire1p and other subunit(s) in the adaptor complex may be required for a productive interaction leading to phosphorylation. Phosphorylation of Gcn5p or other subunit(s) of the Gcn5/Ada complex may target the complex to the UPRE to selectively activate transcription of genes encoding ER chaperons. It is also possible that interaction between Ire1p and Gcn5p may not result in phosphorylation, but rather may recruit the promoter to the nuclear membrane via interaction of the complex with Hac1p or another DNA-binding factor. Once the Gcn5/Ada complex is localized to a promoter, two models have been proposed for its role in transcription (32). The complex may function to open up chromatin subsequently leading the transcriptional apparatus through repressive chromatin structures. Alternatively, the chromatin remodeling SWI/SNF complex (33) may open up chromatin, and Gcn5/Ada complex may function to keep the chromatin open for repeated rounds of transcription.
Acknowledgments
We thank Drs. Roger Brent, Russ Finly, and Erica Golemis for providing reagents for the yeast two-hybrid system, Dr. Peter Walter for plasmid pJC005, Dr. Leonard Guarente for providing yeast strains, Dr. Richard Morimoto for helpful discussion, and Drs. Jack Dixon and Ben Margolis for critically reading the manuscript.
ABBREVIATIONS
- ER
endoplasmic reticulum
- UPR
unfolded protein response
- UPRE
UPR element
- HSR
heat shock response
- HSE
HSR element
- GST
glutathione S-transferase
- HA
influenza hemagglutin epitope
- BD
bromodomain
- X-Gal
5-bromo-4-chloro-3-indolyl β-d-galactoside
References
- 1.Gething M-J, Sambrook J. Nature (London) 1992;355:33–45. doi: 10.1038/355033a0. [DOI] [PubMed] [Google Scholar]
- 2.Mori K, Sant A, Kohno K, Normington K, Gething M-J, Sambrook J. EMBO J. 1992;11:2583–2593. doi: 10.1002/j.1460-2075.1992.tb05323.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chang S C, Erwin A E, Lee A S. Mol Cell Biol. 1989;9:2153–2162. doi: 10.1128/mcb.9.5.2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cox J C, Walter P. Cell. 1996;87:391–404. doi: 10.1016/s0092-8674(00)81360-4. [DOI] [PubMed] [Google Scholar]
- 5.Cox J S, Shamu C E, Walter P. Cell. 1993;73:1197–1206. doi: 10.1016/0092-8674(93)90648-a. [DOI] [PubMed] [Google Scholar]
- 6.Mori K, Ma W, Gething M-J, Sambrook J. Cell. 1993;74:743–756. doi: 10.1016/0092-8674(93)90521-q. [DOI] [PubMed] [Google Scholar]
- 7.Welihinda A A, Kaufman R J. J Biol Chem. 1996;271:18181–18187. doi: 10.1074/jbc.271.30.18181. [DOI] [PubMed] [Google Scholar]
- 8.Shamu C E, Walter P. EMBO J. 1996;15:3028–3039. [PMC free article] [PubMed] [Google Scholar]
- 9.Georgakopoulos T, Thireos G. EMBO J. 1992;11:4145–4152. doi: 10.1002/j.1460-2075.1992.tb05507.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marcus G A, Silverman N, Berger S L, Horiuchi J, Guarente L. EMBO J. 1994;13:4807–4815. doi: 10.1002/j.1460-2075.1994.tb06806.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Horiuchi J, Silverman N, Marcus G A, Guarente L. Mol Cell Biol. 1995;15:1203–1209. doi: 10.1128/mcb.15.3.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marcus G A, Horiuchi J, Silverman N, Guarente L. Mol Cell Biol. 1996;16:3197–3205. doi: 10.1128/mcb.16.6.3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brownell J E, Zhouy J, Ranalli T, Kobayashi R, Edmondson D G, Roth S Y, Allis C D. Cell. 1996;84:843–851. doi: 10.1016/s0092-8674(00)81063-6. [DOI] [PubMed] [Google Scholar]
- 14.Gyuris J, Golemis E, Chertkov H, Brent R. Cell. 1993;75:791–803. doi: 10.1016/0092-8674(93)90498-f. [DOI] [PubMed] [Google Scholar]
- 15.Sherman F, Fink G R, Hicks J. Methods in Yeast Genetics. Plainview, NY: Cold Spring Harbor Lab. Press; 1986. [Google Scholar]
- 16.Wach A, Brachat A, Pohlmann R, Philippsen P. Yeast. 1994;10:1793–1808. doi: 10.1002/yea.320101310. [DOI] [PubMed] [Google Scholar]
- 17.Sikroski R S, Hieter P. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Finley R L, Brent R. Proc Natl Acad Sci USA. 1994;91:12980–12984. doi: 10.1073/pnas.91.26.12980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hill J, Donald K A I, Griffiths D E. Nucleic Acids Res. 1991;19:5791. doi: 10.1093/nar/19.20.5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Williams F E, Varanasi U, Trumbly R J. Mol Cell Biol. 1991;11:3307–3316. doi: 10.1128/mcb.11.6.3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harlow E, Lane D. Antibodies: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1988. [Google Scholar]
- 22.Schmitt M F, Brown T A, Trumpower B L. Nucleic Acids Res. 1990;18:3091–3092. doi: 10.1093/nar/18.10.3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 24.LaMantia M, Miura T, Tachikawa H, Kaplan H A, Lennarz W J, Mizunaga T. Proc Natl Acad Sci USA. 1991;88:4453–4457. doi: 10.1073/pnas.88.10.4453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barlev N A, Candau R, Wang L, Darpino P, Silverman N, Berger S L. J Biol Chem. 1995;270:19337–19344. doi: 10.1074/jbc.270.33.19337. [DOI] [PubMed] [Google Scholar]
- 26.Roberts S M, Winston F. Mol Cell Biol. 1996;16:3206–3213. doi: 10.1128/mcb.16.6.3206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nikawa J-I, Yamashita S. Mol Microbiol. 1992;6:1441–1446. doi: 10.1111/j.1365-2958.1992.tb00864.x. [DOI] [PubMed] [Google Scholar]
- 28.Rougvie A E, Lis J T. Cell. 1988;54:795–804. doi: 10.1016/s0092-8674(88)91087-2. [DOI] [PubMed] [Google Scholar]
- 29.Rasmussen E B, Lis J T. J Mol Biol. 1995;252:522–535. doi: 10.1006/jmbi.1995.0517. [DOI] [PubMed] [Google Scholar]
- 30.Allfrey V, Faulkner R M, Mirsky A E. Proc Natl Acad Sci USA. 1964;51:786–790. doi: 10.1073/pnas.51.5.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Braunstein M, Rose A B, Holmes S G, Allis C D, Broach J R. Genes Dev. 1993;7:593–604. doi: 10.1101/gad.7.4.592. [DOI] [PubMed] [Google Scholar]
- 32.Brownell J E, Allis C D. Curr Opin Genet Dev. 1996;6:176–184. doi: 10.1016/s0959-437x(96)80048-7. [DOI] [PubMed] [Google Scholar]
- 33.Wilson C J, Chao D M, Imbalzano A N, Schnitzler G R, Kingston R E, Young R A. Cell. 1996;84:235–244. doi: 10.1016/s0092-8674(00)80978-2. [DOI] [PubMed] [Google Scholar]