Abstract
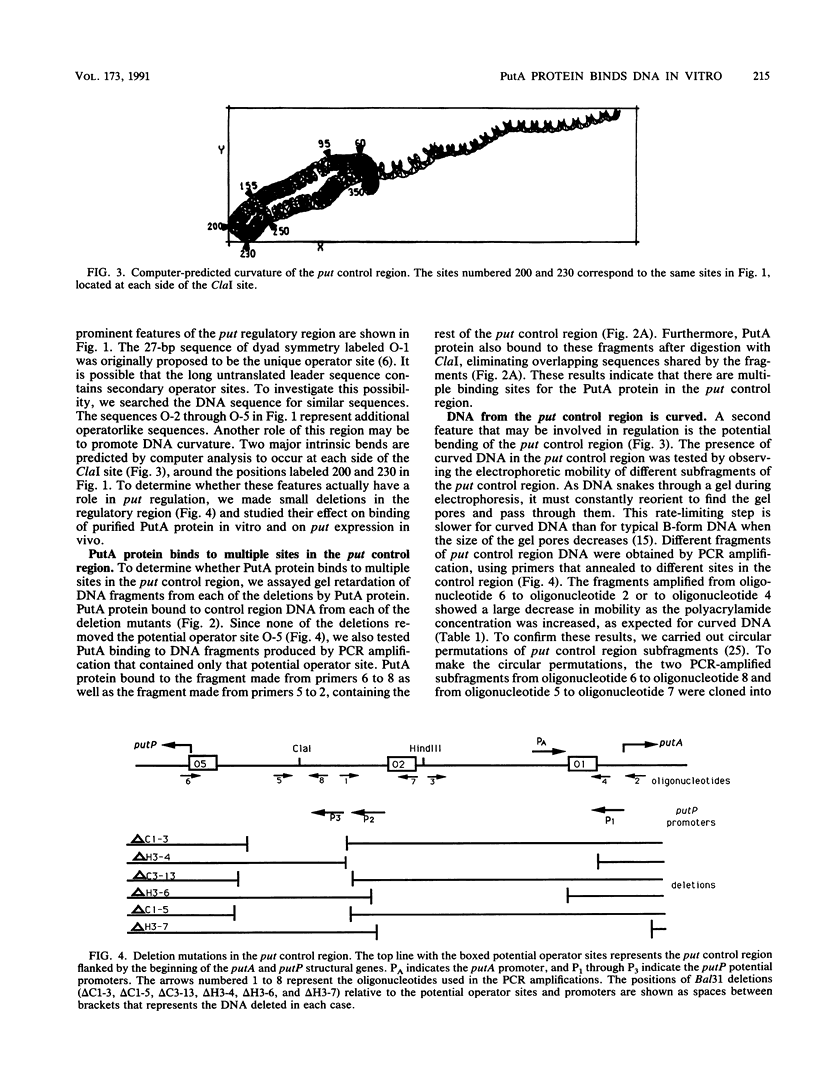
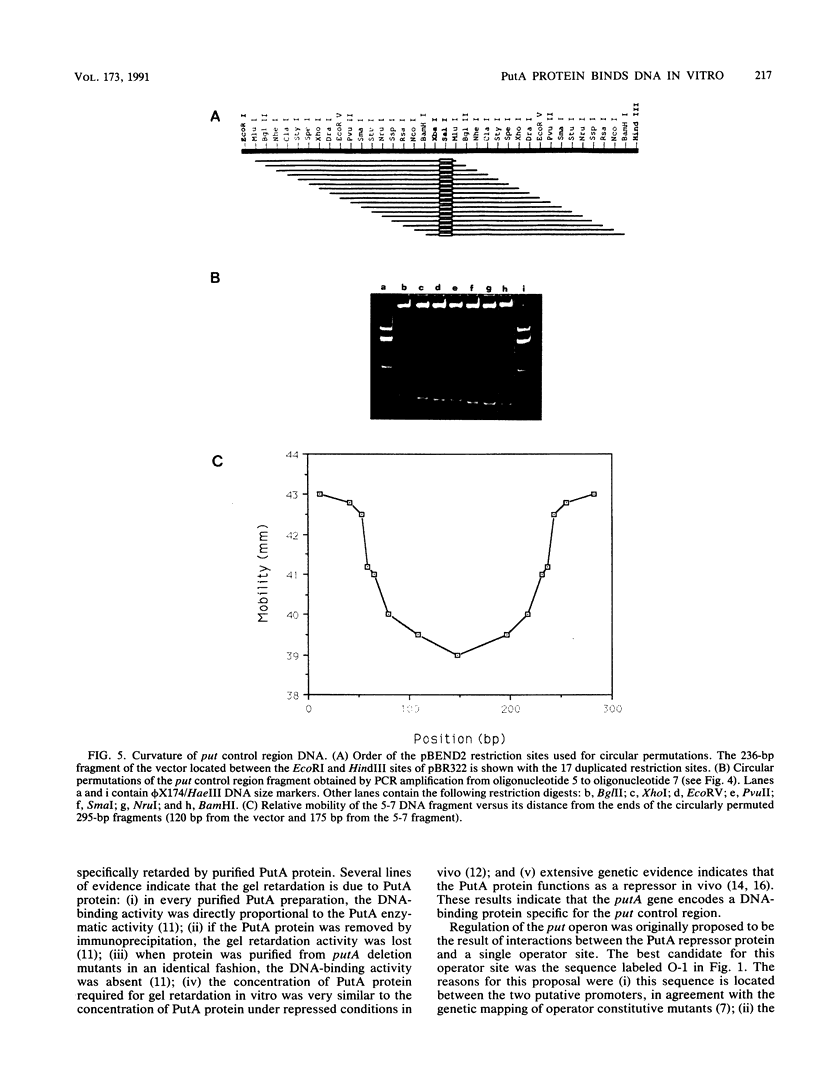
The PutA protein is a membrane-associated enzyme that catalyzes the degradation of proline to glutamate. Genetic evidence suggests that in the absence of proline, the PutA protein also represses transcription of the putA and putP genes. To directly determine whether PutA protein binds to the put control region, we analyzed gel retardation of put control region DNA by purified PutA protein in vitro. The put control region is 420 bp. Purified PutA protein bound specifically to several nonoverlapping fragments of control region DNA, indicating the presence of multiple binding sites in the control region. Electrophoretic abnormalities and behavior of circularly permuted fragments of control region DNA indicate that it contains a region of intrinsically curved DNA. To determine whether the multiple binding sites or the DNA curvature are important in vivo, two types of deletions were constructed: (i) deletions that removed sequences predicted to contribute to DNA curvature as well as potential operator sites and (ii) deletions that removed only potential operator sites. Both types of deletions increased expression of the put genes but were still induced by proline, indicating that multiple cis elements are involved in repression. These data suggest a model for put repression that invokes the formation of a complex between PutA protein molecules bound at different sites in the control region, brought into proximity by a loop of curved DNA.
Full text
PDF
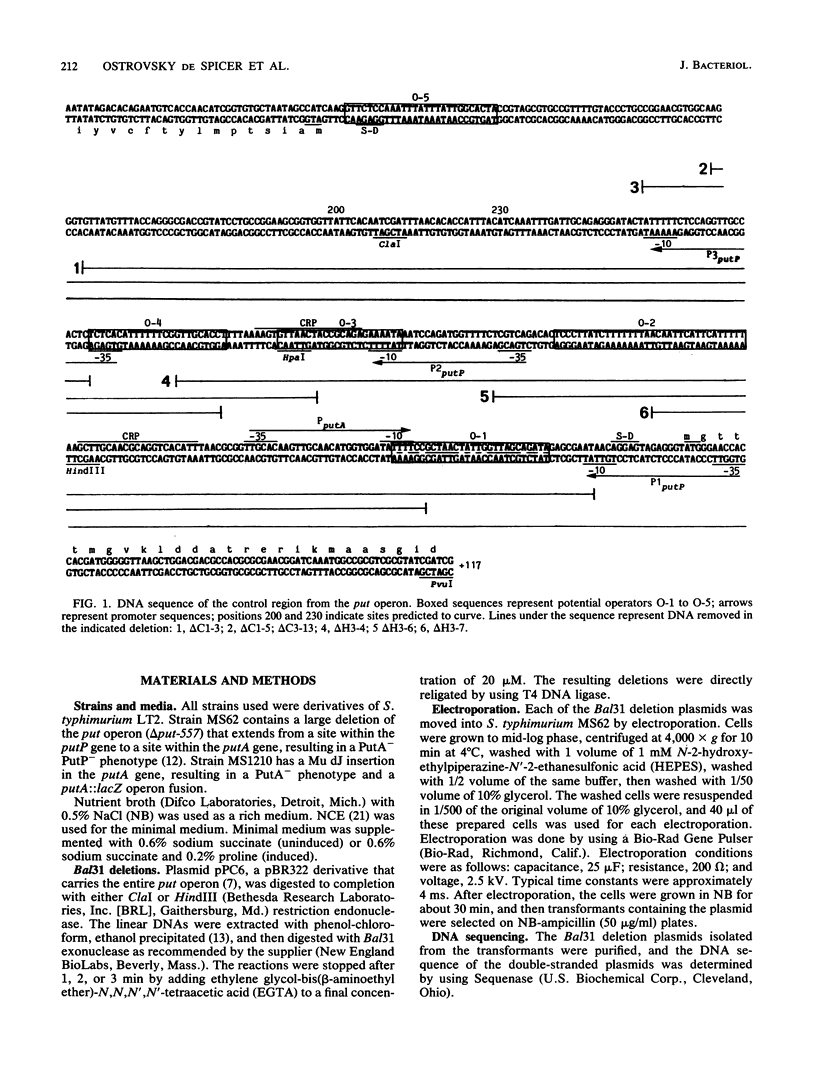

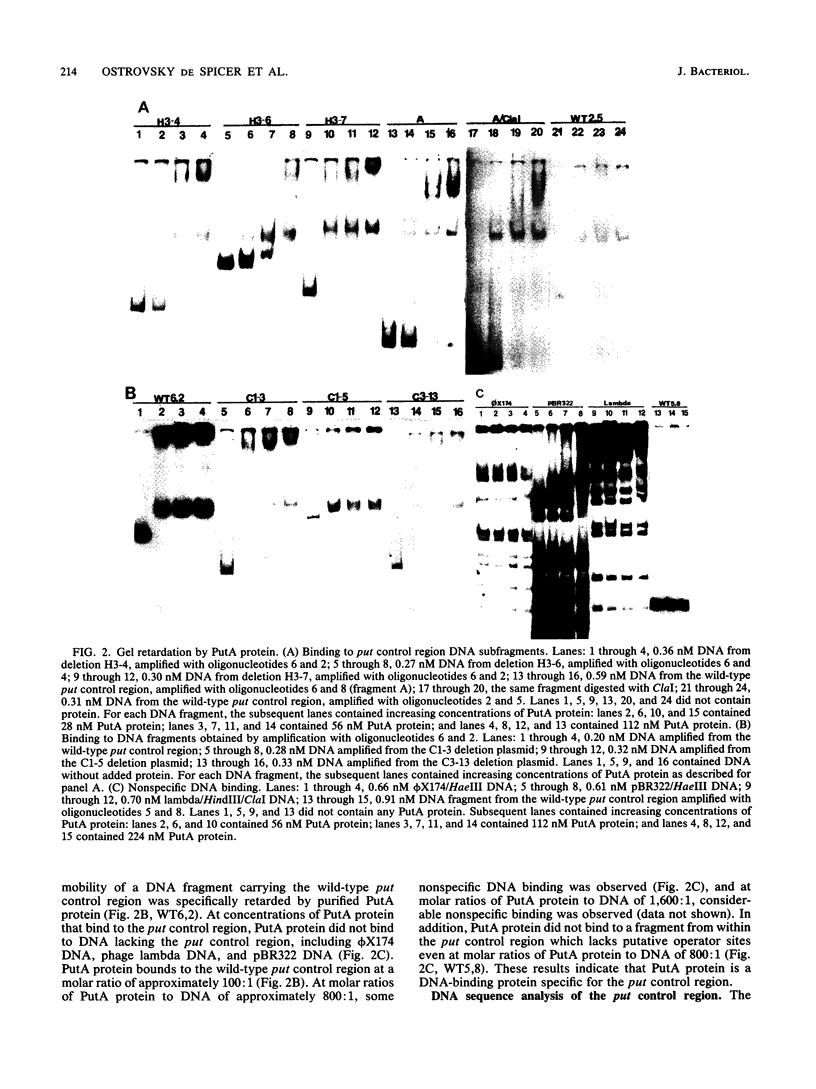



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Dendinger S., Brill W. J. Regulation of proline degradation in Salmonella typhimurium. J Bacteriol. 1970 Jul;103(1):144–152. doi: 10.1128/jb.103.1.144-152.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekena K., Liao M. K., Maloy S. Activation of a new proline transport system in Salmonella typhimurium. J Bacteriol. 1990 Jun;172(6):2940–2945. doi: 10.1128/jb.172.6.2940-2945.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fried M., Crothers D. M. Equilibria and kinetics of lac repressor-operator interactions by polyacrylamide gel electrophoresis. Nucleic Acids Res. 1981 Dec 11;9(23):6505–6525. doi: 10.1093/nar/9.23.6505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garner M. M., Revzin A. A gel electrophoresis method for quantifying the binding of proteins to specific DNA regions: application to components of the Escherichia coli lactose operon regulatory system. Nucleic Acids Res. 1981 Jul 10;9(13):3047–3060. doi: 10.1093/nar/9.13.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn D. R., Maloy S. R. Regulation of the put operon in Salmonella typhimurium: characterization of promoter and operator mutations. Genetics. 1986 Nov;114(3):687–703. doi: 10.1093/genetics/114.3.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn D. R., Myers R. S., Kent C. R., Maloy S. R. Regulation of proline utilization in Salmonella typhimurium: molecular characterization of the put operon, and DNA sequence of the put control region. Mol Gen Genet. 1988 Jul;213(1):125–133. doi: 10.1007/BF00333408. [DOI] [PubMed] [Google Scholar]
- Huo L., Martin K. J., Schleif R. Alternative DNA loops regulate the arabinose operon in Escherichia coli. Proc Natl Acad Sci U S A. 1988 Aug;85(15):5444–5448. doi: 10.1073/pnas.85.15.5444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maloy S. R., Roth J. R. Regulation of proline utilization in Salmonella typhimurium: characterization of put::Mu d(Ap, lac) operon fusions. J Bacteriol. 1983 May;154(2):561–568. doi: 10.1128/jb.154.2.561-568.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marini J. C., Levene S. D., Crothers D. M., Englund P. T. Bent helical structure in kinetoplast DNA. Proc Natl Acad Sci U S A. 1982 Dec;79(24):7664–7668. doi: 10.1073/pnas.79.24.7664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menzel R., Roth J. Purification of the putA gene product. A bifunctional membrane-bound protein from Salmonella typhimurium responsible for the two-step oxidation of proline to glutamate. J Biol Chem. 1981 Sep 25;256(18):9755–9761. [PubMed] [Google Scholar]
- Menzel R., Roth J. Regulation of the genes for proline utilization in Salmonella typhimurium: autogenous repression by the putA gene product. J Mol Biol. 1981 May 5;148(1):21–44. doi: 10.1016/0022-2836(81)90233-3. [DOI] [PubMed] [Google Scholar]
- Morgan B. A., Hayward R. S. Direct evidence for rifampicin-promoted readthrough of the partial terminator tL7 in the rpoBC operon of Escherichia coli. Mol Gen Genet. 1987 Dec;210(2):358–363. doi: 10.1007/BF00325706. [DOI] [PubMed] [Google Scholar]
- Ratzkin B., Roth J. Cluster of genes controlling proline degradation in Salmonella typhimurium. J Bacteriol. 1978 Feb;133(2):744–754. doi: 10.1128/jb.133.2.744-754.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulanovsky L. E., Trifonov E. N. Estimation of wedge components in curved DNA. Nature. 1987 Apr 16;326(6114):720–722. doi: 10.1038/326720a0. [DOI] [PubMed] [Google Scholar]
- Wilbur W. J., Lipman D. J. Rapid similarity searches of nucleic acid and protein data banks. Proc Natl Acad Sci U S A. 1983 Feb;80(3):726–730. doi: 10.1073/pnas.80.3.726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood J. M. Membrane association of proline dehydrogenase in Escherichia coli is redox dependent. Proc Natl Acad Sci U S A. 1987 Jan;84(2):373–377. doi: 10.1073/pnas.84.2.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H. M., Crothers D. M. The locus of sequence-directed and protein-induced DNA bending. Nature. 1984 Apr 5;308(5959):509–513. doi: 10.1038/308509a0. [DOI] [PubMed] [Google Scholar]
- Zwieb C., Kim J., Adhya S. DNA bending by negative regulatory proteins: Gal and Lac repressors. Genes Dev. 1989 May;3(5):606–611. doi: 10.1101/gad.3.5.606. [DOI] [PubMed] [Google Scholar]