Abstract
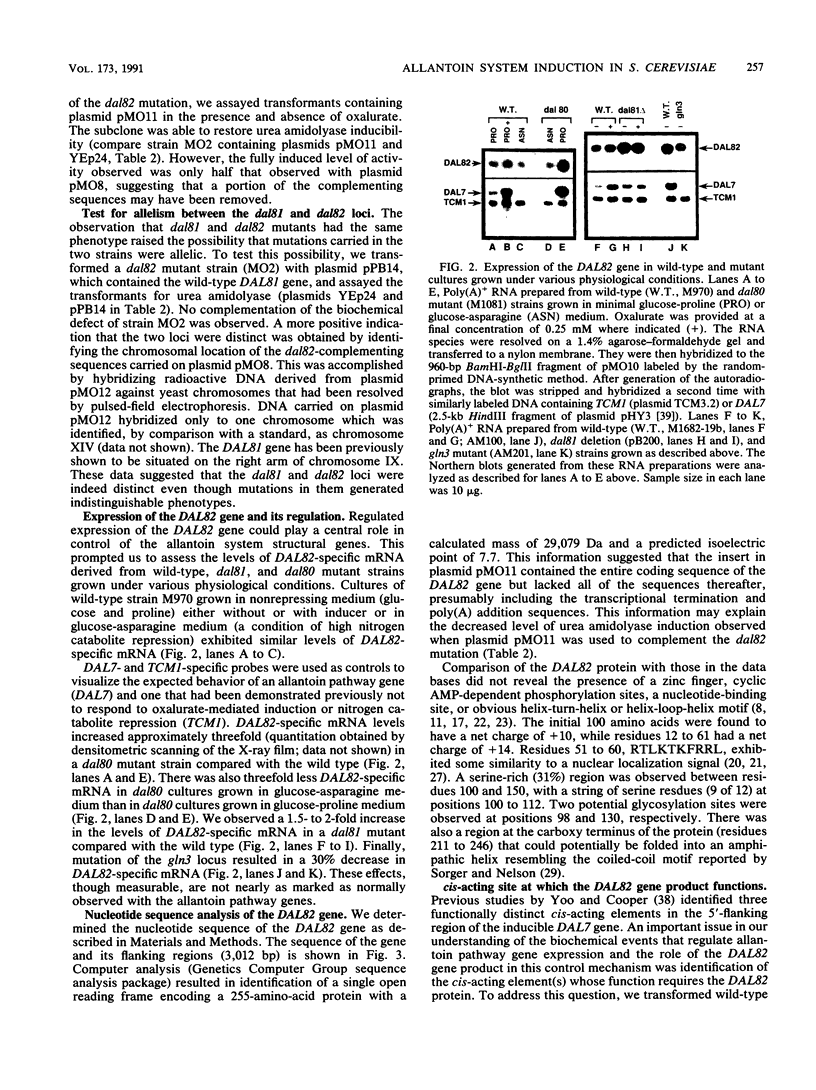
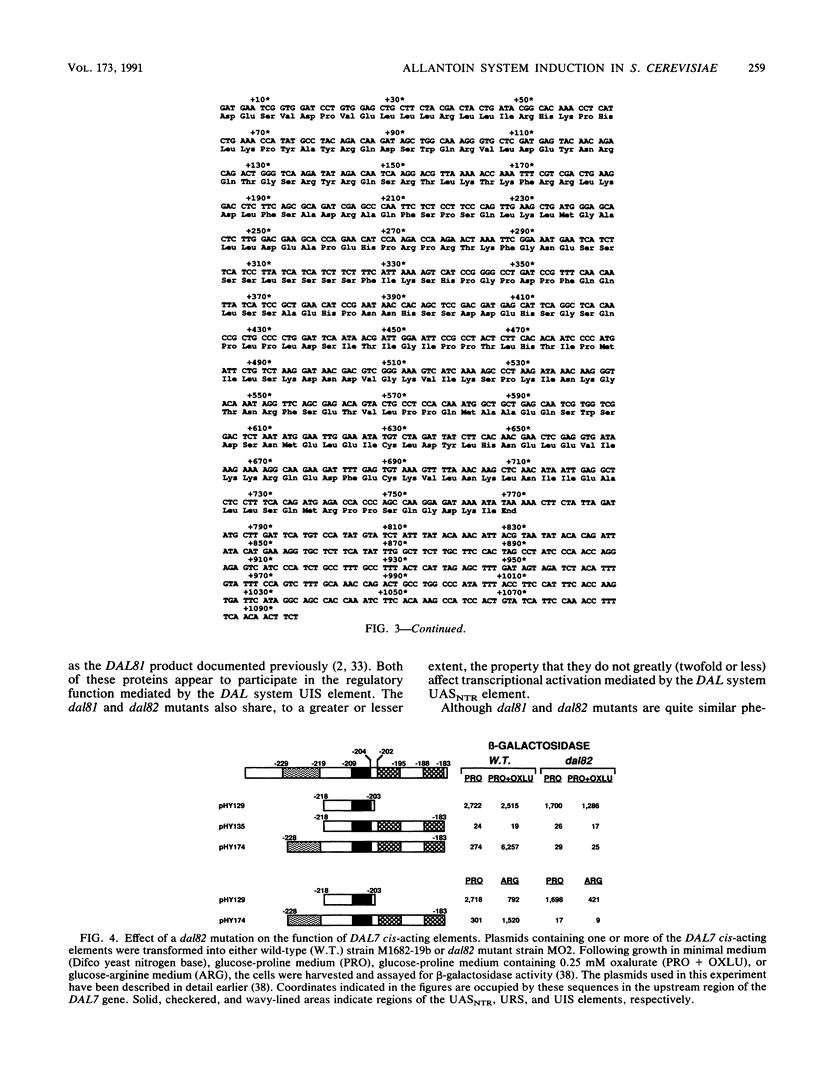
Several highly inducible enzyme activities are required for the degradation of allantoin in Saccharomyces cerevisiae. Induction of these pathway enzymes has been shown to be regulated at transcription, and response to inducer is lost in dal81 and dal82/durM mutants. The similar phenotypes generated by dal81 and dal82 mutations prompted the question of whether they were allelic. We demonstrated that the DAL81 and DAL82 loci are distinct, unlinked genes situated on chromosomes IX and XIV. DAL82 gene expression did not respond to induction by the allantoin pathway inducer or to nitrogen catabolite repression. Expression was also not significantly affected by mutation of the dal80 locus. From the nucleotide sequence of the DAL82 gene, we deduced that it encodes a protein with a mass of 29,079 Da that may possess the structural motifs expected of a regulatory protein. This protein was shown to be required for the function mediated by the cis-acting upstream induction sequence situated in the 5'-flanking regions of the inducible allantoin pathway genes.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baum J. A., Geever R., Giles N. H. Expression of qa-1F activator protein: identification of upstream binding sites in the qa gene cluster and localization of the DNA-binding domain. Mol Cell Biol. 1987 Mar;7(3):1256–1266. doi: 10.1128/mcb.7.3.1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bricmont P. A., Cooper T. G. A gene product needed for induction of allantoin system genes in Saccharomyces cerevisiae but not for their transcriptional activation. Mol Cell Biol. 1989 Sep;9(9):3869–3877. doi: 10.1128/mcb.9.9.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson M., Botstein D. Two differentially regulated mRNAs with different 5' ends encode secreted with intracellular forms of yeast invertase. Cell. 1982 Jan;28(1):145–154. doi: 10.1016/0092-8674(82)90384-1. [DOI] [PubMed] [Google Scholar]
- Cooper T. G., Chisholm V. T., Cho H. J., Yoo H. S. Allantoin transport in Saccharomyces cerevisiae is regulated by two induction systems. J Bacteriol. 1987 Oct;169(10):4660–4667. doi: 10.1128/jb.169.10.4660-4667.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper T. G., Ferguson D., Rai R., Bysani N. The GLN3 gene product is required for transcriptional activation of allantoin system gene expression in Saccharomyces cerevisiae. J Bacteriol. 1990 Feb;172(2):1014–1018. doi: 10.1128/jb.172.2.1014-1018.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper T. G., Rai R., Yoo H. S. Requirement of upstream activation sequences for nitrogen catabolite repression of the allantoin system genes in Saccharomyces cerevisiae. Mol Cell Biol. 1989 Dec;9(12):5440–5444. doi: 10.1128/mcb.9.12.5440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courchesne W. E., Magasanik B. Regulation of nitrogen assimilation in Saccharomyces cerevisiae: roles of the URE2 and GLN3 genes. J Bacteriol. 1988 Feb;170(2):708–713. doi: 10.1128/jb.170.2.708-713.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelman A. M., Blumenthal D. K., Krebs E. G. Protein serine/threonine kinases. Annu Rev Biochem. 1987;56:567–613. doi: 10.1146/annurev.bi.56.070187.003031. [DOI] [PubMed] [Google Scholar]
- Evans T., Felsenfeld G. The erythroid-specific transcription factor Eryf1: a new finger protein. Cell. 1989 Sep 8;58(5):877–885. doi: 10.1016/0092-8674(89)90940-9. [DOI] [PubMed] [Google Scholar]
- Genbauffe F. S., Cooper T. G. Induction and repression of the urea amidolyase gene in Saccharomyces cerevisiae. Mol Cell Biol. 1986 Nov;6(11):3954–3964. doi: 10.1128/mcb.6.11.3954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins C. F., Hiles I. D., Salmond G. P., Gill D. R., Downie J. A., Evans I. J., Holland I. B., Gray L., Buckel S. D., Bell A. W. A family of related ATP-binding subunits coupled to many distinct biological processes in bacteria. Nature. 1986 Oct 2;323(6087):448–450. doi: 10.1038/323448a0. [DOI] [PubMed] [Google Scholar]
- Hinnen A., Hicks J. B., Fink G. R. Transformation of yeast. Proc Natl Acad Sci U S A. 1978 Apr;75(4):1929–1933. doi: 10.1073/pnas.75.4.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman C. S., Winston F. A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene. 1987;57(2-3):267–272. doi: 10.1016/0378-1119(87)90131-4. [DOI] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs E., Dubois E., Wiame J. M. Regulation of ureaamidolyase synthesis in Saccharomyces cerevisiae, RNA analysis, and cloning of the positive regulatory gene DURM. Curr Genet. 1985;9(5):333–339. doi: 10.1007/BF00421602. [DOI] [PubMed] [Google Scholar]
- Johnston M. A model fungal gene regulatory mechanism: the GAL genes of Saccharomyces cerevisiae. Microbiol Rev. 1987 Dec;51(4):458–476. doi: 10.1128/mr.51.4.458-476.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemper B., Jackson P. D., Felsenfeld G. Protein-binding sites within the 5' DNase I-hypersensitive region of the chicken alpha D-globin gene. Mol Cell Biol. 1987 Jun;7(6):2059–2069. doi: 10.1128/mcb.7.6.2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell A. P., Magasanik B. Regulation of glutamine-repressible gene products by the GLN3 function in Saccharomyces cerevisiae. Mol Cell Biol. 1984 Dec;4(12):2758–2766. doi: 10.1128/mcb.4.12.2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreland R. B., Langevin G. L., Singer R. H., Garcea R. L., Hereford L. M. Amino acid sequences that determine the nuclear localization of yeast histone 2B. Mol Cell Biol. 1987 Nov;7(11):4048–4057. doi: 10.1128/mcb.7.11.4048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreland R. B., Nam H. G., Hereford L. M., Fried H. M. Identification of a nuclear localization signal of a yeast ribosomal protein. Proc Natl Acad Sci U S A. 1985 Oct;82(19):6561–6565. doi: 10.1073/pnas.82.19.6561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murre C., McCaw P. S., Baltimore D. A new DNA binding and dimerization motif in immunoglobulin enhancer binding, daughterless, MyoD, and myc proteins. Cell. 1989 Mar 10;56(5):777–783. doi: 10.1016/0092-8674(89)90682-x. [DOI] [PubMed] [Google Scholar]
- Pabo C. O., Sauer R. T. Protein-DNA recognition. Annu Rev Biochem. 1984;53:293–321. doi: 10.1146/annurev.bi.53.070184.001453. [DOI] [PubMed] [Google Scholar]
- Plumb M., Frampton J., Wainwright H., Walker M., Macleod K., Goodwin G., Harrison P. GATAAG; a cis-control region binding an erythroid-specific nuclear factor with a role in globin and non-globin gene expression. Nucleic Acids Res. 1989 Jan 11;17(1):73–92. doi: 10.1093/nar/17.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai R., Genbauffe F. S., Sumrada R. A., Cooper T. G. Identification of sequences responsible for transcriptional activation of the allantoate permease gene in Saccharomyces cerevisiae. Mol Cell Biol. 1989 Feb;9(2):602–608. doi: 10.1128/mcb.9.2.602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai R., Genbauffe F., Lea H. Z., Cooper T. G. Transcriptional regulation of the DAL5 gene in Saccharomyces cerevisiae. J Bacteriol. 1987 Aug;169(8):3521–3524. doi: 10.1128/jb.169.8.3521-3524.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee S. K., Icho T., Wickner R. B. Structure and nuclear localization signal of the SKI3 antiviral protein of Saccharomyces cerevisiae. Yeast. 1989 May-Jun;5(3):149–158. doi: 10.1002/yea.320050304. [DOI] [PubMed] [Google Scholar]
- Rose M. D., Novick P., Thomas J. H., Botstein D., Fink G. R. A Saccharomyces cerevisiae genomic plasmid bank based on a centromere-containing shuttle vector. Gene. 1987;60(2-3):237–243. doi: 10.1016/0378-1119(87)90232-0. [DOI] [PubMed] [Google Scholar]
- Sorger P. K., Nelson H. C. Trimerization of a yeast transcriptional activator via a coiled-coil motif. Cell. 1989 Dec 1;59(5):807–813. doi: 10.1016/0092-8674(89)90604-1. [DOI] [PubMed] [Google Scholar]
- Sumrada R. A., Cooper T. G. Isolation of the CAR1 gene from Saccharomyces cerevisiae and analysis of its expression. Mol Cell Biol. 1982 Dec;2(12):1514–1523. doi: 10.1128/mcb.2.12.1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumrada R., Cooper T. G. Oxaluric acid: a non-metabolizable inducer of the allantoin degradative enzymes in Saccharomyces cerevisiae. J Bacteriol. 1974 Mar;117(3):1240–1247. doi: 10.1128/jb.117.3.1240-1247.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai S. F., Martin D. I., Zon L. I., D'Andrea A. D., Wong G. G., Orkin S. H. Cloning of cDNA for the major DNA-binding protein of the erythroid lineage through expression in mammalian cells. Nature. 1989 Jun 8;339(6224):446–451. doi: 10.1038/339446a0. [DOI] [PubMed] [Google Scholar]
- Turoscy V., Chisholm G., Cooper T. G. Location of the genes that control induction of the allantoin-degrading enzymes in Saccharomyces cerevisiae. Genetics. 1984 Dec;108(4):827–831. doi: 10.1093/genetics/108.4.827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turoscy V., Cooper T. G. Pleiotropic control of five eucaryotic genes by multiple regulatory elements. J Bacteriol. 1982 Sep;151(3):1237–1246. doi: 10.1128/jb.151.3.1237-1246.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West B. L., Catanzaro D. F., Mellon S. H., Cattini P. A., Baxter J. D., Reudelhuber T. L. Interaction of a tissue-specific factor with an essential rat growth hormone gene promoter element. Mol Cell Biol. 1987 Mar;7(3):1193–1197. doi: 10.1128/mcb.7.3.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitney P. A., Cooper T. G., Magasanik B. The induction of urea carboxylase and allophanate hydrolase in Saccharomyces cerevisiae. J Biol Chem. 1973 Sep 10;248(17):6203–6209. [PubMed] [Google Scholar]
- Wickerham L. J. A Critical Evaluation of the Nitrogen Assimilation Tests Commonly Used in the Classification of Yeasts. J Bacteriol. 1946 Sep;52(3):293–301. [PMC free article] [PubMed] [Google Scholar]
- Yoo H. S., Cooper T. G. The DAL7 promoter consists of multiple elements that cooperatively mediate regulation of the gene's expression. Mol Cell Biol. 1989 Aug;9(8):3231–3243. doi: 10.1128/mcb.9.8.3231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo H. S., Genbauffe F. S., Cooper T. G. Identification of the ureidoglycolate hydrolase gene in the DAL gene cluster of Saccharomyces cerevisiae. Mol Cell Biol. 1985 Sep;5(9):2279–2288. doi: 10.1128/mcb.5.9.2279. [DOI] [PMC free article] [PubMed] [Google Scholar]




