Abstract
Fatty acids (FAs) and their derivatives are essential cellular metabolites whose concentrations must be closely regulated. This implies that regulatory circuits exist which can sense changes in FA levels. Indeed, the peroxisome proliferator-activated receptor α (PPARα) regulates lipid homeostasis and is transcriptionally activated by a variety of lipid-like compounds. It remains unclear as to how these structurally diverse compounds can activate a single receptor. We have developed a novel conformation-based assay that screens activators for their ability to bind to PPARα/δ and induce DNA binding. We show here that specific FAs, eicosanoids, and hypolipidemic drugs are ligands for PPARα or PPARδ. Because altered FA levels are associated with obesity, atherosclerosis, hypertension, and diabetes, PPARs may serve as molecular sensors that are central to the development and treatment of these metabolic disorders.
Fatty acids (FAs) are ubiquitous biological molecules that are used as metabolic fuels, as covalent regulators of signaling molecules, and as essential components of cellular membranes. It is thus logical that FA levels should be closely regulated. Indeed, some of the most common medical disorders in industrialized societies (cardiovascular disease, hyperlipidemia, obesity, and insulin resistance) are characterized by altered levels of FAs or their metabolites (1, 2).
The need for precise control of FA levels suggests that organisms possess sensors that can respond to changes in the available levels of FA metabolites. Peroxisome proliferator-activated receptor α (PPARα) has been identified as a vertebrate nuclear hormone receptor which regulates genes involved in FA degradation (β- and ω-oxidation) (3). PPARα is highly expressed in the liver and was originally identified by Issemann and Green (4) as a molecule that mediates the transcriptional effects of drugs that induce peroxisome proliferation in rodents. Mice lacking functional PPARα are incapable of responding to these agents and fail to induce expression of a variety of genes required for the metabolism of FAs in peroxisomes, mitochondria, and other cellular compartments (5). As a result, PPARα-deficient mice inappropriately accumulate lipid in response to pharmacologic stimuli.
PPARα is a member of the nuclear receptor superfamily that includes receptors for the steroid, thyroid, and retinoid hormones (6). Two other PPARα-related genes (PPARγ and PPARδ) have been identified in mammals. PPARγ is highly enriched in adipocytes while the δ isoform is ubiquitously expressed (3). Like other members of this superfamily, PPARs contain a central DNA-binding domain that recognizes response elements in the promoters of their target genes. PPAR response elements (PPREs) are composed of a directly repeating core-site separated by 1 nt (7). To recognize a PPRE, PPARs must heterodimerize with the 9-cis-retinoic acid receptor (RXR).
Once bound to a response element, PPARs activate transcription through a conserved C-terminal ligand binding domain. Although no ligand has been identified for PPARα, sequence analysis indicates that its C-terminal region is similar to the ligand binding domains of known nuclear hormone receptors. This has prompted an intense search for the identification of ligands for the PPARs. Recently, we and others (8, 9) have identified 15-deoxy-Δ12,14-prostaglandin J2 (15d-J2) as a ligand for PPARγ. Activation of PPARγ by 15d-J2 or its synthetic analogs (thiazolidinediones) (8) promotes differentiation of pre-adipocytes into mature, triglyceride-containing fat cells. Similarly, thiazolidinediones have been shown to increase body weight in animals (10), suggesting that 15d-J2 may be used as an in vivo signal to store FAs in the form of triglycerides.
In contrast to the γ isoform, PPARα appears to regulate FA oxidation, suggesting that PPARα ligands may represent endogenous signals for FA degradation (3). Issemann and Green (4) originally demonstrated that PPARα is activated by fibrates, a group of drugs that induce peroxisome proliferation and FA oxidation in rodents. These drugs are currently being used as serum triglyceride lowering agents. Because fibrates and polyunsaturated FAs (PUFAs) were known to possess similar activities, Gottlicher et al. (11) examined the ability of FAs to activate PPARα. These studies and others have uncovered a bewildering array of compounds (Fig. 1A) that can activate PPARα (3). However, all attempts to demonstrate that these compounds bind directly to PPARα have failed. This has led to the suggestion that these compounds alter FA metabolism which indirectly leads to the accumulation of an endogenous PPARα ligand (13). We have developed a novel ligand-binding assay that facilitates the identification of ligands for PPARα and PPARδ. Contrary to common belief, we find that fibrates and specific FAs/eicosanoids can bind to these receptors. This indicates that FAs simultaneously serve as intermediary metabolites and as primary regulators of transcriptional networks. In addition, the demonstration of a direct interaction between fibrates and PPARα suggests that this receptor could be used as a target for the rapid identification of highly potent and selective hypolipidemic agents.
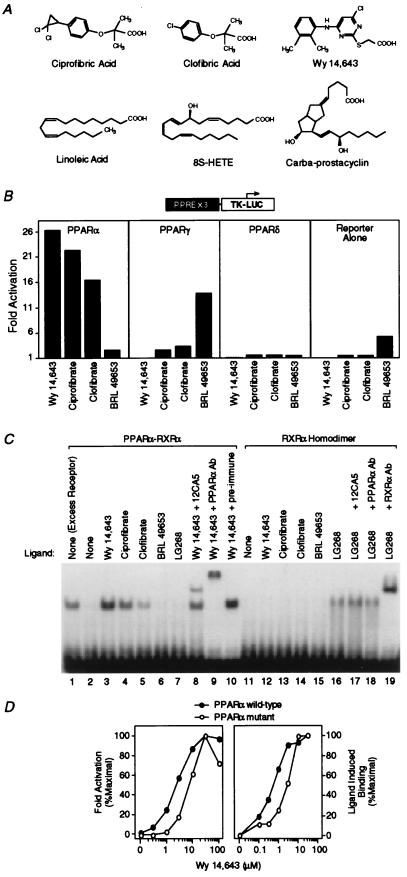
Figure 1.
Hypolipidemic fibrates are ligands for PPARα. (A) Chemical structures of some compounds that we demonstrate to be ligands for PPARα or -δ. (B) Fibrates selectively activate PPARα in a cell-based transient transfection assay. Cells were treated with the following concentrations of each compound: 5 μM Wy 14,643, 300 μM ciprofibrate, 300 μM clofibrate, and 1 μM BRL 49653. (C) Fibrates selectively promote binding of PPARα–RXRα heterodimers to labeled DNA in an electrophoretic mobility shift assay. Compounds were added at the following concentrations: 5 μM Wy 14,643, 100 μM ciprofibrate, 1,000 μM clofibrate, 1 μM BRL 49653, and 1 μM LG268. In lane 1 (excess receptor), the amounts of PPARα and RXRα were increased to 0.6 μl and 0.5 μl, respectively. Where indicated, 1 μl of antibody was added to the reaction. (D) Comparison of the dose response profile of wild-type PPARα with PPARα-G (Glu-282 → Gly) (12) in the transient transfection assay (Left) and the LIC assay (Right). The ligand-induced complex was quantified by phosphorimaging analysis. Ligand-induced binding represents the amount of complex produced at any concentration of ligand minus that produced in the absence of ligand. Maximal induced binding was defined to be 100%; binding observed at other concentrations was normalized to this value.
MATERIALS AND METHODS
Cell Culture and Transfection.
CV-1 cells were grown and transfected as described (8). The reporter construct, PPREx3 TK-LUC contained three copies of the acyl-CoA oxidase PPRE upstream of the Herpes virus thymidine kinase promoter (7). Expression vectors contained the cytomegalovirus IE promoter/enhancer (pCMX) upstream of wild-type mouse PPARα, mouse PPARγ1-ΔN (Met-105–Tyr-475), mouse PPARδ-ΔN (Leu-69–Tyr-440), mouse PPARα-G (Glu-282 → Gly) (12), or Escherichia coli β-galactosidase as an internal control. Cells were exposed to the compounds for 24 h and then harvested and assayed for luciferase and β-galactosidase activity. All points were performed in triplicate and varied by less than 10%. Normalized luciferase activity was determined and plotted as fold-activation relative to untreated cells. Each experiment was repeated three or more times with similar results.
Electrophoretic Mobility-Shift Assays.
In vitro-translated mouse PPARα (0.2 μl) and human RXRα (0.1 μl) were incubated for 30 min at room temperature with 100,000 cpm of Klenow-labeled acyl-CoA oxidase PPRE as described (14) but with 150 mM KCl.
RESULTS
Hypolipidemic Drugs Are PPARα Ligands.
To evaluate the selectivity of PPARs toward hypolipidemic drugs, CV-1 cells were transiently transfected with a PPAR-responsive reporter, PPAR expression vectors, and then treated with various hypolipidemic agents (Fig. 1B). Wy 14,643 and BRL 49653 were included as positive controls because these compounds selectively activate PPARα and -γ, respectively (8, 9, 15). The hypolipidemic fibrates ciprofibrate and clofibrate activated PPARα maximally at 300 μM and exhibited only weak activity on PPARγ (Fig. 1B). Similar results were seen with gemfibrozil (data not shown). In contrast, at 1 mM, the effective serum concentration of clofibrate (16), all three drugs displayed significant activity (5- to 9-fold) on PPARγ (data not shown). These compounds are ineffective activators of PPARδ (Fig. 1B), suggesting that hypolipidemic activity is mediated by PPARα and perhaps by PPARγ.
We sought to determine whether these compounds are PPARα ligands. In the past, classical ligand binding assays have been used to identify ligands for other nuclear receptors. This approach has not been informative in the case of PPARα because radiolabeled ligands are either not available or produce unacceptable levels of nonspecific binding. To overcome these limitations, we developed an assay that does not use a labeled ligand. Our approach relies on the ability of nuclear receptor ligands to induce conformational changes that promote dimerization and subsequent DNA binding. In previous studies, ecdysone (17), vitamin D (18), and 9-cis-retinoic acid (19) were shown to enhance the dimerization and DNA binding activities of their respective receptors. Accordingly, we examined whether PPARα activators could induce similar events. Previous mobility shift assays have demonstrated that PPARα–RXR heterodimers bind to PPREs as obligate heterodimers even in the absence of ligand (15). Indeed, using standard conditions in which both receptors are in excess, PPARα-RXRα heterodimers are readily observed (Fig. 1C, lane 1). However, when both receptors are limiting, binding activity is minimal (Fig. 1C, lane 2) but is dramatically enhanced by Wy 14,643, ciprofibric, or clofibric acids (Fig. 1C, lanes 3–5). This enhancement is unique to PPARα activators as enhanced binding was not observed with PPARγ-specific ligands such as BRL 49653 (Fig. 1C, lane 6), pioglitazone, and troglitazone (data not shown) or the RXR-specific ligands LG268 (Fig. 1C, lane 7), LG69, and 9-cis-retinoic acid (data not shown). PPARα and RXRα are components of the ligand-induced complex since it is supershifted by PPARα-specific (Fig. 1C, lane 9) and RXRα-specific antibodies (data not shown) but not by pre-immune serum (Fig. 1C, lane 10). Similarly, epitope-tagged PPARα is supershifted by an epitope-specific mAb (12CA5) (Fig. 1C, lane 8). Control experiments indicate that PPAR activators do not promote the DNA binding activity of an RXR homodimer (Fig. 1C, lanes 11–15), which is inducible by RXR-specific ligands (Fig. 1C, lanes 16–19). These experiments suggest that ligand-induced complex formation (LIC) represents a sensitive approach for the identification of novel ligands for orphan nuclear receptors.
To further validate the LIC assay, we compared the dose-response profiles of wild-type PPARα to that of a previously characterized point mutant (PPARα-G) (12) that exhibits a decreased potency for PPARα activators in cotransfection experiments. As expected, the concentration required for half-maximal transcriptional activation by Wy 14,643 was 4-fold greater with the mutant receptor (Fig. 1D Left). In the LIC assay, phosphorimaging analysis revealed a similar increase in the amount of Wy 14,643 required for half-maximal ligand-induced binding (LIC50) with the mutant receptor (Fig. 1D Right). Thus, the LIC50 for Wy 14,643 (600 nM) appears to provide an effective estimate of the actual dissociation constant. These data both confirm the validity of the LIC assay and provide evidence that hypolipidemic agents such as Wy 14,643, ciprofibrate, and clofibrate are direct ligands for PPARα.
Long-Chain FAs Are PPARα Ligands.
We utilized the LIC assay to determine which, if any, naturally occurring FAs bind to PPARα at physiologic concentrations. In the fasting state, the total concentration of nonesterified FAs in serum is ≈700 μM (20). Abundant dietary FAs such as linoleic and arachidonic acid have average concentrations of 25–30 μM and may reach much higher levels (Richard Wilkinson, HyClone, personal communication). The intracellular concentrations of these compounds are more difficult to determine but can be inferred from the Michaelis constant of long-chain fatty-acyl-CoA synthetase (21) (LC-FACS, 20 μM). Thus, we examined the ability of a variety of FAs to activate PPARα at 30 μM concentrations. When compared with Wy 14,643 in the cotransfection assay, saturated short-chain FAs (<C10) were poor activators of PPARα while longer-chain FAs (C10-C16) possessed weak activity (Fig. 2A). Surprisingly, 30 μM doses of long-chain FAs (≥C12) induced complex formation in the LIC assay (Fig. 2B). A carboxyl group is required for this activity since the corresponding fatty alcohols neither activated nor induced binding (Fig. 2 A and B). These data indicate that long-chain FAs can bind weakly to PPARα.
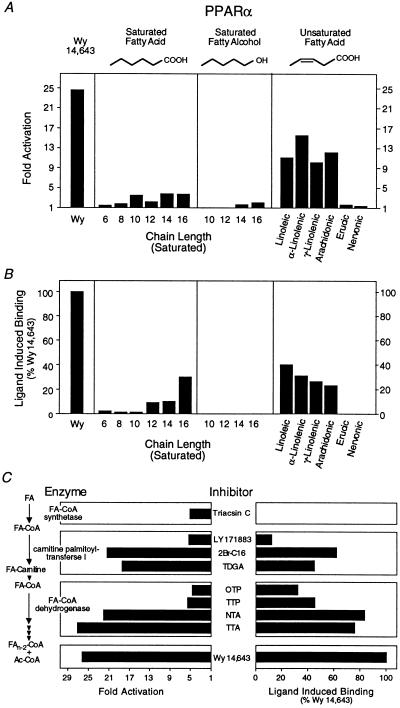
Figure 2.
Long-chain FAs and β-oxidation inhibitors are PPARα ligands. (A) Activation of PPARα by FAs and fatty alcohols. All compounds were added to a final concentration of 30 μM except for Wy 14,643, which was used at 5 μM. (B) Enhancement of PPARα–RXRα heterodimer formation by FAs and fatty alcohols. All compounds were added to a final concentration of 30 μM except for Wy 14,643, which was added to a final concentration of 5 μM. Saturated FAs and fatty alcohols are indicated by their chain length. Unsaturated FAs are as follows: linoleic (cis-Δ9,12-C18:2), α-linolenic (cis-Δ9,12,15-C18:3), γ-linolenic (cis-Δ6,9,12-C18:3), arachidonic (cis-Δ5,8,11,14-C20:4), erucic (cis-Δ13-C22:1), and nervonic (cis-Δ15-C24:1) acids. (C) Inhibitors of β-oxidation activation (Left) and bind (Right) to PPARα. Experiments were performed as described in Fig. 1. Triacsin C (10 μM, Left; 30 μM, Right) was used as an inhibitor of fatty LC-FACS. Inhibitors of carnitine palmitoyltransferase I included LY 171883 (30 μM), 2Br-C16 (5 μM), and tetradecylglycidic acid (TDGA, 5 μM). Fatty-acyl-CoA dehydrogenase was inhibited with octylthioproprionic acid (OTP, 30 μM), tetradecylthioproprionic acid (TTP, 30 μM), nonylthioacetic acid (NTA, 30 μM), and tetradecylthioacetic acid (TTA, 30 μM). Wy 14,643 (5 μM) was included as a positive control.
We next examined the ability of PUFAs to bind to PPARα. We found that linoleic, α-linolenic, γ-linolenic, arachidonic (Fig. 2 A and B, Right), docosahexaenoic, and eicosapentaenoic acids (data not shown) all bound to and activated PPARα. In contrast, very-long-chain unsaturated FAs such as erucic and nervonic acids failed to bind or activate PPARα (Fig. 2 A and B, Right). This structure-activity relationship suggests that PPARα ligands can be broadly defined as long-chain monocarboxylic acids. Optimal binding activity is observed with compounds containing a 16–20 carbon chain length with several double bonds in the chain.
Dual-Function PPARα Activators.
The structural requirements for PPARα binding are reminiscent of the substrate specificity previously defined for LC-FACS (21), an intracellular enzyme that converts free FAs to their corresponding acyl-CoA thioesters. In addition to long-chain FAs, several hypolipidemic drugs are also converted to their acyl-CoA thioesters (22–24). Accordingly, we examined the ligand binding properties of several long-chain FA-CoA thioesters and found that they were incapable of inducing binding in the LIC assay (data not shown). This is consistent with the observation that a free carboxyl group is required for recognition by PPARα (Fig. 2 A and B) and suggests that LC-FACS may inactivate PPARα ligands (25). To test this possibility, we assayed the transcriptional activity of PPARα in cells treated with triacsin C, an inhibitor of LC-FACS (26). Surprisingly, we found that triacsin C itself activated PPARα (Fig. 2C Left) but failed to induce PPARα binding in the LIC assay (Fig. 2C Right). These observations are consistent with the hypothesis that inhibition of LC-FACS leads to the accumulation of an endogenous PPARα activator.
LC-FACS catalyzes the first step in the mitochondrial β-oxidation cascade (Fig. 2C Left). Several groups have shown that inhibitors of subsequent steps in this pathway lead to activation of PPARα and peroxisome proliferation (13, 27, 28). This has contributed to the “lipid-overload” hypothesis which suggests that these inhibitors activate PPARα by promoting the accumulation of an endogenous ligand. However, because these enzymatic inhibitors are structural analogs of long-chain FAs, we addressed the possibility that they might also be PPARα ligands. Consistent with previous results, inhibitors of carnitine palmitoyltransferase I [LY 171883, 2-bromopalmitate (2Br-C16), tetradecylglycidic acid (TDGA)] (29–31) and fatty-acyl-CoA dehydrogenase [octylthioproprionic acid (OTP), tetradecylthioproprionic acid (TTP), nonylthioacetic acid (NTA), tetradecylthioacetic acid (TTA)] (32) all activated PPARα (Fig. 2C Left). Surprisingly, the transcriptional activity of these peroxisome proliferators correlated with their ability to bind PPARα (Fig. 2C Right). Thus, these compounds represent dual-function activators. As ligands they activate PPARα directly; as metabolic inhibitors they may indirectly lead to the accumulation of endogenous FA ligands.
PPARs Are Nuclear Eicosanoid Receptors.
Our data indicate that long-chain FAs bind to PPARα at physiologic concentrations. Because these intermediary metabolites serve as precursors to additional regulators, we wondered whether downstream metabolites may also serve as PPARα ligands. This line of thinking was prompted by our recent demonstration that the arachidonic acid metabolite 15d-J2 is a ligand for the γ isoform of PPAR (8). Accordingly, we asked whether other eicosanoids may be high affinity ligands for PPARα (Fig. 3A). Previous studies (refs. 33–35; B.M.F. and R.M.E, unpublished data) have shown that a number of prostanoids can activate PPARα (Fig. 3A, left). Importantly, when examined in the LIC assay, prostaglandin (PG) I2 analogs such as carbaprostacyclin (cPGI) and iloprost act as ligands while cicaprost (36), a related analog, is inactive (Fig. 3A Right). Thus, agonists for the cell-surface PGI2 receptor exhibit a distinct pharmacologic hierarchy on PPARα. Furthermore, since CV-1 cells lack detectable levels of the PGI2 receptor (34), it appears that this cell-surface pathway is not contributing to PPARα activation.
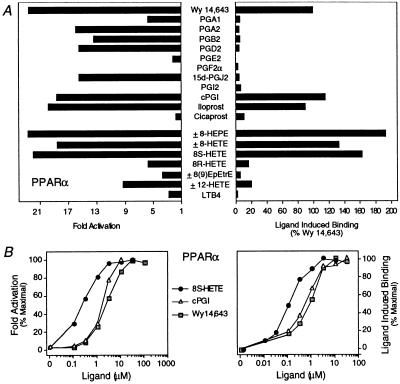
Figure 3.
Identification of eicosanoid ligands for PPARα. (A) cPGI, iloprost, 8(S)-HETE, and 8(S)-HEPE transactivate (Left) and bind (Right) to PPARα. For transfections (Left), compounds were added to cells at the following concentrations: 5 μM Wy 14,643; 10 μM PGA1, PGA2, PGB2, PGD2, PGE2, and PGF2α; 3 μM 15d-J2; 10 μM PGI2; 1 μM cPGI and iloprost; 10 μM cicaprost; 10 μM ±8-HEPE (±8-hydroxy-Δ5Z,9E,11Z,14Z,17Z-C20:5), ±8-HETE (±8-hydroxy-Δ5Z,9E,11Z,14Z-C20:4), ±8(9)-EpEtrE [±8(9)-epoxy-Δ5Z,11Z,14Z-C20:3], and ± 12-HETE (±12-hydroxy-Δ5Z,8Z,10E,14Z-C20:4); 5 μM 8(S)- and 8(R)-HETE; and 10 μM LTB4. For the ligand binding assay (Right), compounds were added as follows: 10 μM Wy 14,643, PGA1, PGA2, PGB2, PGD2, PGE2, PGF2α, 15d-J2, and PGI2; 2 μM cPGI, iloprost, and cicaprost; 1 μM ±8-HEPE, ±8-HETE, ±8(9)-EpEtrE, and ±12-HETE; 300 nM 8(S)-HETE and 8(R)-HETE; and 10 μM LTB4. (B) Dose-response curves comparing the potency of 8(S)-HETE, cPGI, and Wy 14,643 in transactivating (Left) and binding (Right) to PPARα.
In searching for additional eicosanoid ligands, we focused our attention on oxygenated FA derivatives and other products of lipoxygenase metabolism. While leukotriene B4 (LTB4) (37) and other lipoxygenase products were poor or ineffective ligands (Fig. 3 and data not shown), 8(S)-hydroxyeicosatetraenoic acid [8(S)-HETE] was, as previously reported (33), an effective activator of PPARα (Fig. 3A Left). Further structure-activity studies revealed that ±8-hydroxyeicosatrienoic acid (±8-HETrE) was significantly less effective (data not shown) whereas ±8-hydroxyeicosapentaenoic acid (±8-HEPE) was a slightly more effective activator (Fig. 3A Left). When examined in the LIC assay, ±8-HETE and ± 8-HEPE both served as PPARα ligands (Fig. 3A Right). The stereochemistry around the 8-position was crucial since 8(R)-HETE was a poor ligand and a poor activator of PPARα (Fig. 3A). Dose-response studies (Fig. 3B) revealed that 8(S)-HETE and cPGI activate with half-maximal activity at 200 nM and 2 μM, respectively (Fig. 3B Left) and bind PPARα with affinities estimated to be 100 nM and 500 nM, respectively (Fig. 3B Right). Thus, the naturally occurring 8(S)-HETE is the highest affinity ligand yet to be identified for PPARα.
The data in Fig. 3A indicate that certain compounds can activate PPARα without inducing complex formation in vitro. This could occur if these compounds represented inactive precursors that are metabolized to ligands. Alternatively, they could bind to PPARα without inducing a conformation change that promotes DNA binding. To rule out this possibility, PPARα–RXRα heterodimers were formed in the presence of Wy 14,643, and an excess of each compound that failed to induce complex formation. A compound that binds to PPARα without inducing complex formation would be expected to compete with Wy 14,643 thereby decreasing heterodimer formation. All of the compounds tested (LTB4, BRL 49653, PGA1, PGA2, PGB2, PGD2, PGE2, PGF2α, PGI2, 15d-J2, and cicaprost) were ineffective inhibitors of Wy 14,643-enhanced binding (data not shown), suggesting that these compounds are not ligands for PPARα. Thus, PPARα activators such as PGA1, PGA2, PGB2, PGD2, and 15d-J2 may be inactive precursors that are metabolized to PPARα ligands.
PPARα and -δ Possess Overlapping Ligand Specificities.
Because ligands have not been discovered for PPARδ, we wondered whether FAs or eicosanoids may also bind to this receptor. At concentrations that were sufficient for activation of PPARα, a number of hypolipidemic agents, thiazolidinediones, and saturated FAs failed to bind or activate PPARδ (Figs. 1B and 4A and data not shown). In contrast, several PUFAs and eicosanoids activated PPARδ (Fig. 4A Left) and a subset of these (linoleic acid, arachidonic acid, cPGI, and iloprost) acted as ligands in the LIC assay (Fig. 4A Right). Taken together, our data indicate that the PPARs comprise a family of nuclear FA and eicosanoid receptors.
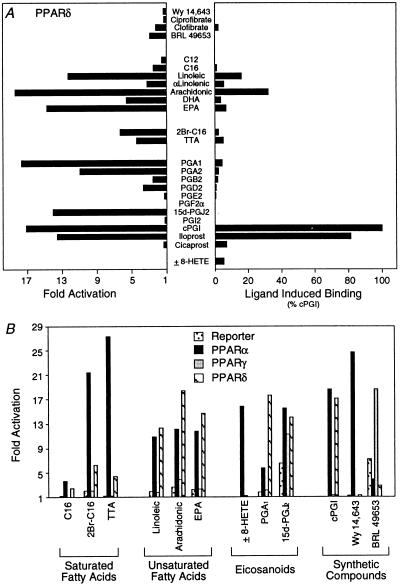
Figure 4.
PPARα/δ and -γ display distinct ligand response profiles. (A) Linoelic acid, arachidonic acid, cPGI, and iloprost transactivate (Left) and bind (Right) to PPARδ. After transfection (Left), compounds were added to cells at the following concentrations: 5 μM Wy 14,643; 100 μM ciprofibrate; 1,000 μM clofibrate; 5 μM BRL 49653; 30 μM C12, C16, linoleic acid, α-linoleic, arachidonic, docosahexaenoic (DHA, all-Z-Δ4,7,10,13,16,19-C22:6), and eicosapentaenoic (EPA, all-Z-Δ5,8,11,14,17-C20:5) acids; 5 μM 2Br-C16; 30 μM TTA; 10 μM PGA1, PGA2, PGB2, PGD2, PGE2, and PGF2α; 3 μM 15d-J2; 10 μM PGI2; 1 μM cPGI and iloprost; 10 μM cicaprost; and 3 μM ±8-HETE. For the ligand binding assay (Right), compounds were added as follows: 5 μM Wy 14,643; 100 μM ciprofibrate; 1,000 μM clofibrate; 50 μM BRL 49653; 30 μM C12, C16, linoleic acid, α-linoleic, arachidonic acids, DHA, and EPA; 10 μM 2Br-C16, TTA, PGA1, PGA2, PGB2, PGD2, PGE2, PGF2α, 15d-J2, PGI2, cPGI, iloprost, and cicaprost; and 1 μM ±8-HETE. (B) Comparison of the responsiveness of PPARα, -γ, and -δ to various compounds. After transfection, cells were treated with the following concentrations of compounds: 30 μM C16; 5 μM 2Br-C16; 30 μM TTA, linoleic, arachidonic acids, and EPA; 3 μM ±8-HETE; 10 μM PGA1; 3 μM 15d-J2; 1 μM cPGI; and 5 μM Wy 14,643 and BRL 49653.
Finally, we compared the specificity of different activator classes for each member of the PPAR family (Fig. 4B and data not shown). Naturally occurring saturated long-chain FAs (C12–C16) are weak activators of PPARα and even weaker activators of PPARδ. The dual function long-chain FAs (2Br-C16, TTA) preferentially activate PPARα over PPARδ. In contrast, PUFAs are efficient activators of PPARα and -δ, but display little activity on PPARγ. Among the eicosanoids, 8(S)-HETE was specific for PPARα, while PGA1 preferentially activated PPARδ (33). All three PPAR isoforms were responsive to 15d-J2 whereas the synthetic eicosanoid cPGI selectively activated PPARα and -δ. These data indicate that PPARα, -γ, and -δ are a family of nuclear receptors that possess distinct, yet overlapping, ligand binding specificities.
DISCUSSION
Metabolite-Mediated Transcriptional Control.
The identification of mammalian nuclear receptors with FA and eicosanoid ligands have a number of important implications. First, this establishes an important link between metabolism and transcriptional control. PPARα induces transcription of a number of gene products that contribute to the metabolism of FAs. These include enzymes necessary for the degradation of FAs through β- and ω-oxidation pathways. It has long been established that metabolic intermediates modulate feedback control by promoting allosteric changes in enzymatic activity. The demonstration that FAs bind to PPARα provides direct evidence that metabolic intermediates can also regulate transcription. This complements the immediate effects of allosteric control by modulating the metabolic capacities of the organism over longer time periods. Transcriptional control by metabolic intermediates has long been appreciated in bacteria and yeast. For example, the lac and trp repressors coordinately regulate transcription by binding to micromolar concentrations of allolactose and tryptophan (38, 39), respectively. Similarities between the lac operon and PPARα-regulated transcription are particularly striking. In both cases, metabolic precursors (lactose/FAs) are converted to higher affinity inducers [allolactose/8(S)-HETE] that coordinately regulate the synthesis of enzymes required for the catabolism of the initial metabolites (lactose/FAs). Our data strongly suggest that metabolite-controlled intracellular (metacrine) signaling systems are operative in higher organisms. The development of the LIC assay may facilitate the identification of other metacrine signals that function as micromolar ligands for other orphan nuclear receptors.
We have shown that PPARα can recognize a broad array of ligands. This is unique among the nuclear receptors and suggests that PPARα senses broad changes in FA status and dietary inputs. In particular, as metabolism may vary from cell-to-cell and tissue-to-tissue, PPARα may act locally to integrate a variety of cell-specific metabolic parameters. In contrast to PPARα which promotes FA catabolism, PPARγ appears to stimulate the opposing function of FA storage. We show that PPARα ligands are distinct from those of PPARγ. The ability of these receptors to respond to distinct metabolic cues provides a potential mechanism for the animal to maintain a balance between FA breakdown and storage. Although a function for PPARδ remains to be established, it is of interest to note that this receptor recognizes a subset of PPARα ligands suggesting that it may respond to similar endogenous signals. Thus, the overall balance between FA catabolism and storage may be determined by the relative levels PPARα/δ and PPARγ ligands.
We have shown that 8(S)-HETE is a high affinity ligand for PPARα. It is unclear what function this ligand has, however, its identification in the skin (40, 41) suggests that it may play a specialized function in this tissue. In contrast to 8(S)-HETE, other eicosanoids were found that activate but fail to bind to PPAR (e.g., PGA1 and PPARδ) (33) (Figs. 3A and 4A). By analogy to all-trans-retinoic acid which binds to RXR after conversion to the active ligand (9-cis-retinoic acid) (6), these eicosanoids may represent precursors to additional PPAR ligands. Thus, additional eicosanoid ligands may exist and their production could be regulated in a tissue-specific manner.
A previous report suggested that LTB4 binds Xenopus PPARα with an affinity of ≈100 nM (37). However, nonspecific binding to PPARα was not accounted for and half-maximal displacement required 10–50 μM of unlabeled LTB4. Because we were unable to detect activation or binding with 10 μM LTB4 (Fig. 3A), it is unclear whether LTB4 is a physiologically relevant ligand for mouse PPARα.
PPAR, Dietary FAs, and Human Disease.
The ability to regulate FA pools is essential for normal homeostasis. Indeed, inappropriately high levels of triglycerides and nonesterified FAs are a common component of obesity, insulin resistance, hypertension, and hyperlipidemia (1, 2). These abnormalities often develop in the same individual and are ominous signs of impending coronary heart disease, a major cause of death in industrialized societies. It has been proposed that increased levels of triglycerides and FAs are key factors in the progression of these disorders, which suggests that normalization of these parameters could contribute to an effective therapy. Indeed, it is well known that dietary PUFAs can be beneficial in this regard (42, 43). This may reflect both activation of PPAR-regulated β- and ω-oxidation pathways (44) as well as PUFA-dependent suppression of lipogenic and glycolytic enzymes (45). A negative PUFA response element has been identified in the promoter of the pyruvate kinase gene (46). This response element binds HNF-4, a constitutively active orphan nuclear receptor whose DNA-binding specificity overlaps that of PPAR–RXR heterodimers. Other investigators have shown that PPARα antagonizes HNF-4 by down-regulating its expression in liver and by binding nonproductively to HNF-4 response elements (47). These observations, along with our demonstration that PUFAs promote the binding of PPARα/δ–RXRα heterodimers, suggest that PUFAs may suppress transcription by displacing constitutively active HNF-4 and replacing it with an abortive PPARα/δ–RXRα complex. Thus, in addition to promoting β- and ω-oxidation, PPARα and -δ may also inhibit lipogenesis. Taken together, these observations suggest that PPARα and -δ may directly mediate some of the beneficial effects of dietary PUFAs.
In addition to dietary factors, drugs of the fibrate class are also known to regulate transcription of apolipoproteins A-I, A-II, and C-III (3) and are useful for the treatment of hyperlipidemias. However, the effective doses of the best available drugs are in the high micromolar range. Our demonstration that fibrates bind directly to PPARα suggests that screening for high affinity PPARα ligands may provide a rapid approach for the development of more effective treatments for these lipid-related disorders. Because PPAR isoforms have distinct functions, the relative specificity of a drug for each PPAR isoform may be an important factor in evaluating its therapeutic potential.
In conclusion, our findings suggest that PPARs play a central role in a signaling system that controls lipid homeostasis in higher organisms. As the number of orphan receptors continue to grow, it is likely that these proteins will provide important tools for the discovery of additional regulatory signals.
Acknowledgments
We thank the following investigators for generous contributions of reagents: Eric Johnson, Jon Bremer, Satoshi Omura, Masakazu Hirata, and R. W. Johnson. This work was supported by the Howard Hughes Medical Institute (R.M.E.), the March of Dimes (R.M.E.), the Mathers Foundation, and the Tobacco-Related Disease Research Program (B.M.F.). R.M.E. is an Investigator of the Howard Hughes Medical Institute at the Salk Institute for Biological Studies.
ABBREVIATIONS
- FA
fatty acid
- PPAR
peroxisome proliferator-activated receptor
- PPRE
PPAR response element
- RXR
9-cis-retinoic acid receptor
- 15d-J2
15-deoxy-Δ12,14-prostaglandin J2
- PUFA
polyunsaturated fatty acid
- LIC
ligand-induced complex formation
- LC-FACS
long-chain fatty-acyl-CoA synthetase
- 2Br-C16
2-bromopalmitate
- TTA
tetradecylthioacetic acid
- cPGI
carbaprostacyclin
- LTB4
leukotriene B4
- 8-HETE
8-hydroxyeicosatetraenoic acid
- 8-HEPE
8-hydroxyeicosapentaenoic
- PG
prostaglandin
References
- 1.Durrington, P. N. (1993) Postgrad. Med. J. 69, (Suppl 1), S18–S25; discussion S25–S29. [PubMed]
- 2.Reaven G M. J Intern Med Suppl. 1994;736:13–22. [PubMed] [Google Scholar]
- 3.Schoonjans K, Staels B, Auwerx J. Biochim Biophys Acta. 1996;1302:93–109. doi: 10.1016/0005-2760(96)00066-5. [DOI] [PubMed] [Google Scholar]
- 4.Issemann I, Green S. Nature (London) 1990;347:645–650. doi: 10.1038/347645a0. [DOI] [PubMed] [Google Scholar]
- 5.Lee S S T, Pineau T, Drago J, Lee E J, Owens J W, Kroetz D L, Fernandez-Salguero P M, Westphal H, Gonzalez F J. Mol Cell Biol. 1995;15:3012–3022. doi: 10.1128/mcb.15.6.3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mangelsdorf D J, Evans R M. Cell. 1995;83:841–850. doi: 10.1016/0092-8674(95)90200-7. [DOI] [PubMed] [Google Scholar]
- 7.Kliewer S A, Umesono K, Noonan D J, Heyman R A, Evans R M. Nature (London) 1992;358:771–774. doi: 10.1038/358771a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Forman B M, Tontonoz P, Chen J, Brun R P, Spiegelman B M, Evans R M. Cell. 1995;83:803–812. doi: 10.1016/0092-8674(95)90193-0. [DOI] [PubMed] [Google Scholar]
- 9.Kliewer S A, Lenhard J M, Willson T M, Patel I, Morris D C, Lehmann J M. Cell. 1995;83:813–819. doi: 10.1016/0092-8674(95)90194-9. [DOI] [PubMed] [Google Scholar]
- 10.Zhang B, Graziano M P, Doebber T W, Leibowitz M D, White-Carrington S, Szalkowski D M, Hey P J, Wu M, Cullinan C A, Bailey P, Lollmann B, Frederich R, Flier J S, Strader C D, Smith R G. J Biol Chem. 1996;271:9455–9459. doi: 10.1074/jbc.271.16.9455. [DOI] [PubMed] [Google Scholar]
- 11.Gottlicher M, Widmark E, Li Q, Gustafsson J A. Proc Natl Acad Sci USA. 1992;89:4653–4657. doi: 10.1073/pnas.89.10.4653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hsu M H, Palmer C N, Griffin K J, Johnson E F. Mol Pharmacol. 1995;48:559–567. [PubMed] [Google Scholar]
- 13.Gottlicher M, Demoz A, Svensson D, Tollet P, Berge R K, Gustafsson J A. Biochem Pharmacol. 1993;46:2177–2184. doi: 10.1016/0006-2952(93)90607-x. [DOI] [PubMed] [Google Scholar]
- 14.Forman B M, Goode E, Chen J, Oro A E, Bradley D J, Perlmann T, Noonan D J, Burka L T, McMorris T, Lamph W W, Evans R M, Weinberger C. Cell. 1995;81:687–693. doi: 10.1016/0092-8674(95)90530-8. [DOI] [PubMed] [Google Scholar]
- 15.Kliewer S A, Forman B M, Blumberg B, Ong E S, Borgmeyer U, Mangelsdorf D J, Umesono K, Evans R M. Proc Natl Acad Sci USA. 1994;91:7355–7359. doi: 10.1073/pnas.91.15.7355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Havel R J, Kane J P. Annu Rev Pharmacol. 1973;13:287–308. doi: 10.1146/annurev.pa.13.040173.001443. [DOI] [PubMed] [Google Scholar]
- 17.Yao T P, Forman B M, Jiang Z, Cherbas L, Chen J D, McKeown M, Cherbas P, Evans R M. Nature (London) 1993;366:476–479. doi: 10.1038/366476a0. [DOI] [PubMed] [Google Scholar]
- 18.Cheskis B, Freedman L P. Mol Cell Biol. 1994;14:3329–3338. doi: 10.1128/mcb.14.5.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang X K, Lehmann J, Hoffmann B, Dawson M I, Cameron J, Graupner G, Hermann T, Tran P, Pfahl M. Nature (London) 1992;358:587–591. doi: 10.1038/358587a0. [DOI] [PubMed] [Google Scholar]
- 20.Groop L C, Bonadonna R C, Simonson D C, Petrides A S, Shank M, DeFronzo R A. Am J Physiol. 1992;263:E79–E84. doi: 10.1152/ajpendo.1992.263.1.E79. [DOI] [PubMed] [Google Scholar]
- 21.Tanaka T, Hosaka K, Hoshimaru M, Numa S. Eur J Biochem. 1979;98:165–172. doi: 10.1111/j.1432-1033.1979.tb13173.x. [DOI] [PubMed] [Google Scholar]
- 22.Bronfman M, Morales M N, Amigo L, Orellana A, Nunez L, Cardenas L, Hidalgo P C. Biochem J. 1992;284:289–295. doi: 10.1042/bj2840289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aarsland A, Berge R K. Biochem Pharmacol. 1991;41:53–61. doi: 10.1016/0006-2952(91)90010-3. [DOI] [PubMed] [Google Scholar]
- 24.Wu P, Bremer J. Biochim Biophys Acta. 1994;1215:87–92. doi: 10.1016/0005-2760(94)90095-7. [DOI] [PubMed] [Google Scholar]
- 25.Hertz R, Berman I, Bar-Tana J. Eur J Biochem. 1994;221:611–615. doi: 10.1111/j.1432-1033.1994.tb18773.x. [DOI] [PubMed] [Google Scholar]
- 26.Tomoda H, Igarashi K, Omura S. Biochim Biophys Acta. 1987;921:595–598. [PubMed] [Google Scholar]
- 27.Gulick T, Cresci S, Caira T, Moore D D, Kelly D P. Proc Natl Acad Sci USA. 1994;91:11012–11016. doi: 10.1073/pnas.91.23.11012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Asiedu D K, Skorve J, Willumsen N, Demoz A, Berge R K. Biochim Biophys Acta. 1993;1166:73–76. doi: 10.1016/0005-2760(93)90285-h. [DOI] [PubMed] [Google Scholar]
- 29.Foxworthy P S, Eacho P I. Biochem J. 1988;252:409–414. doi: 10.1042/bj2520409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brady P S, Dunker A K, Brady L J. Biochem J. 1987;241:751–757. doi: 10.1042/bj2410751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kiorpes T C, Hoerr D, Ho W, Weaner L E, Inman M G, Tutwiler G F. J Biol Chem. 1984;259:9750–9755. [PubMed] [Google Scholar]
- 32.Hovik R, Osmundsen H, Berge R, Aarsland A, Bergseth S, Bremer J. Biochem J. 1990;270:167–173. doi: 10.1042/bj2700167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yu K, Bayona W, Kallen C B, Harding H P, Ravera C P, McMahon G, Brown M, Lazar M A. J Biol Chem. 1995;270:23975–23983. doi: 10.1074/jbc.270.41.23975. [DOI] [PubMed] [Google Scholar]
- 34.Hertz R, Berman I, Keppler D, Bar-Tana J. Eur J Biochem. 1996;235:242–247. doi: 10.1111/j.1432-1033.1996.00242.x. [DOI] [PubMed] [Google Scholar]
- 35.Brun R P, Tontonoz P, Forman B M, Ellis R, Chen J, Evans R M, Spiegelman B M. Genes Dev. 1996;10:974–984. doi: 10.1101/gad.10.8.974. [DOI] [PubMed] [Google Scholar]
- 36.Namba T, Oida H, Sugimoto Y, Kakizuka A, Negishi M, Ichikawa A, Narumiya S. J Biol Chem. 1994;269:9986–9992. [PubMed] [Google Scholar]
- 37.Devchand P R, Keller H, Peters J M, Vazquez M, Gonzalez F J, Wahli W. Nature (London) 1996;384:39–43. doi: 10.1038/384039a0. [DOI] [PubMed] [Google Scholar]
- 38.Jobe A, Bourgeois S. J Mol Biol. 1972;69:397–408. doi: 10.1016/0022-2836(72)90253-7. [DOI] [PubMed] [Google Scholar]
- 39.He J J, Matthews K S. J Biol Chem. 1990;265:731–737. [PubMed] [Google Scholar]
- 40.Furstenberger G, Hagedorn H, Jacobi T, Besemfelder E, Stephan M, Lehmann W D, Marks F. J Biol Chem. 1991;266:15738–15745. [PubMed] [Google Scholar]
- 41.Hughes M A, Brash A R. Biochim Biophys Acta. 1991;1081:347–354. doi: 10.1016/0005-2760(91)90292-p. [DOI] [PubMed] [Google Scholar]
- 42.Willumsen N, Skorve J, Hexeberg S, Rustan A C, Berge R K. Lipids. 1993;28:683–690. doi: 10.1007/BF02535987. [DOI] [PubMed] [Google Scholar]
- 43.Spady D K, Woollett L A, Dietschy J M. Annu Rev Nutr. 1993;13:355–381. doi: 10.1146/annurev.nu.13.070193.002035. [DOI] [PubMed] [Google Scholar]
- 44.Green S. Mutat Res. 1995;333:101–109. doi: 10.1016/0027-5107(95)00136-0. [DOI] [PubMed] [Google Scholar]
- 45.Jump D B, Clarke S D, Thelen A, Liimatta M. J Lipid Res. 1994;35:1076–1084. [PubMed] [Google Scholar]
- 46.Liimatta M, Towle H C, Clarke S, Jump D B. Mol Endocrinol. 1994;8:1147–1153. doi: 10.1210/mend.8.9.7838147. [DOI] [PubMed] [Google Scholar]
- 47.Hertz R, Seckbach M, Zakin M M, Bar-Tana J. J Biol Chem. 1996;271:218–224. doi: 10.1074/jbc.271.1.218. [DOI] [PubMed] [Google Scholar]