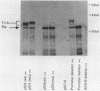
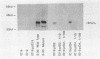
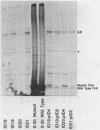
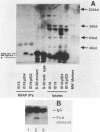
Abstract
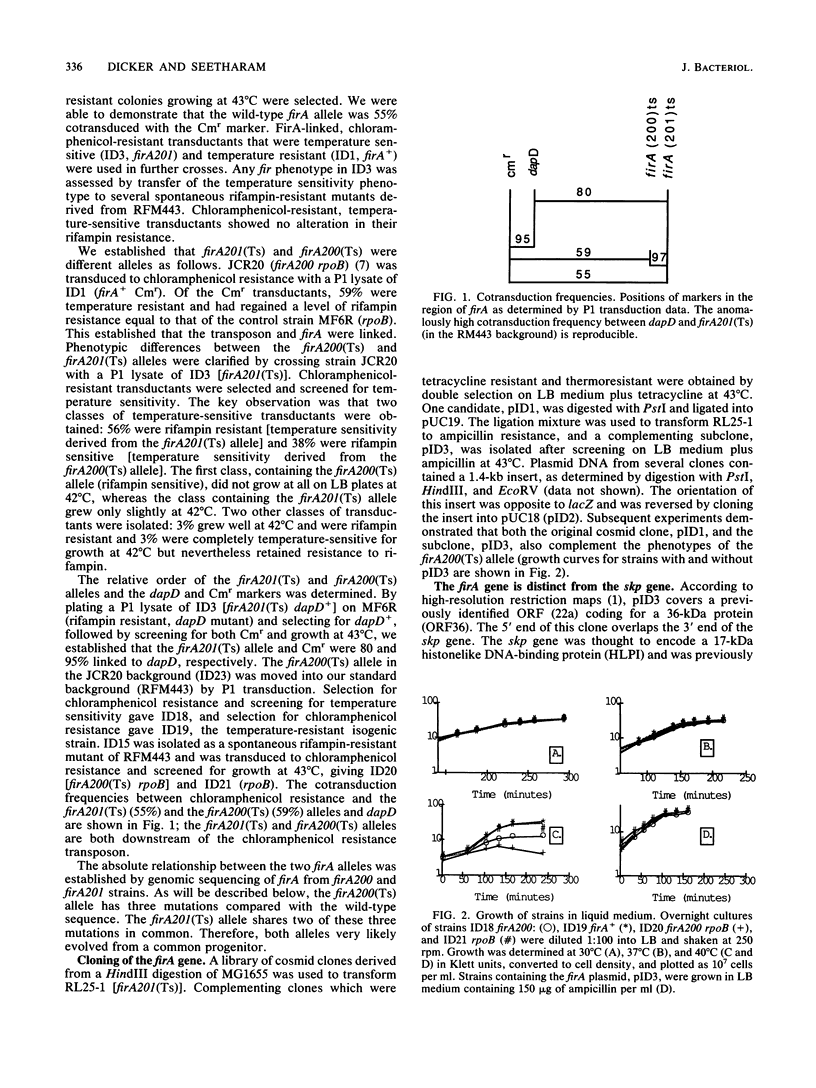
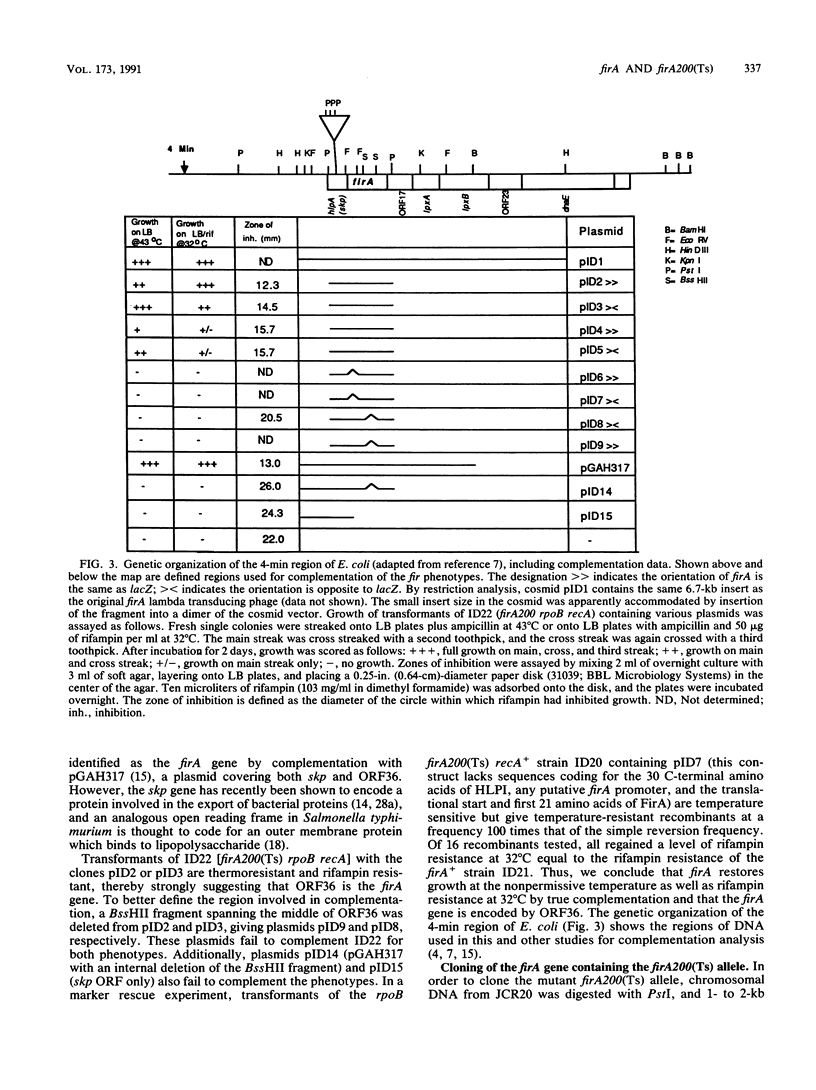
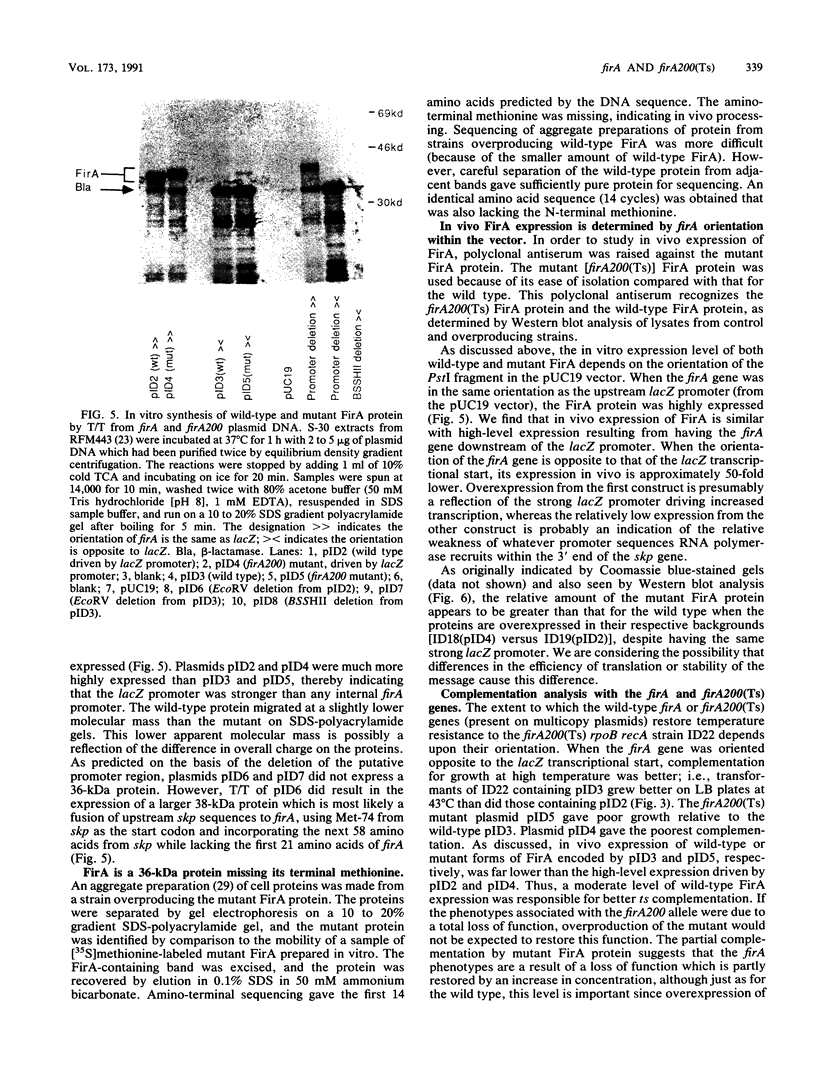
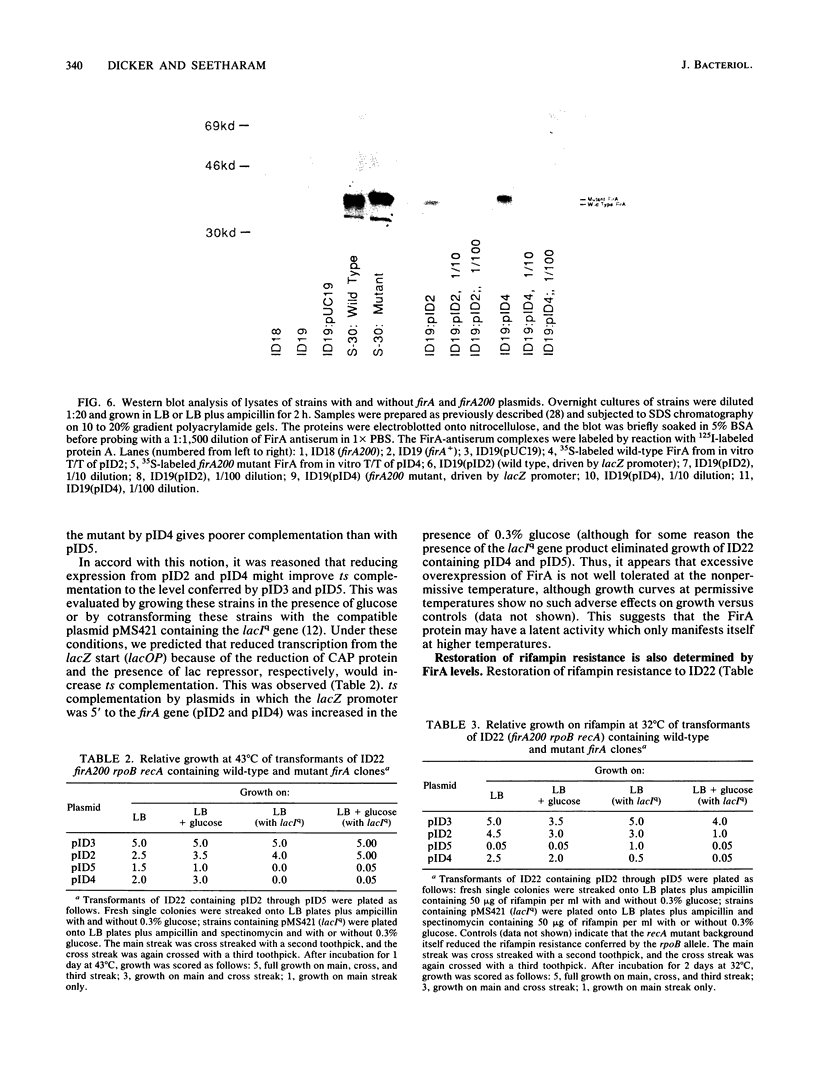
The Escherichia coli gene firA, previously reported to code for a small, histonelike DNA-binding protein, has been cloned and found to reside immediately downstream from skp, a gene previously identified as the firA locus. firA encodes a 36-kDa protein. The mutant firA200(Ts) allele was also cloned and shown to contain three mutations, each mutation giving rise to a single amino acid change. Partially purified wild-type FirA (from a firA+ strain) and mutant FirA [from a firA200(Ts) strain] proteins have amino-terminal sequences predicted from their common DNA sequences. Both proteins lack an N-terminal methionine. Modest overexpression of wild-type or mutant FirA restored wild-type growth to firA200(Ts) strains at 43 degrees C, whereas high-level expression of wild-type FirA was required for more complete suppression of the rifampin sensitivity of firA200(Ts) rpoB double mutants. High-level expression of mutant FirA did not suppress this rifampin sensitivity.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aasland R., Coleman J., Holck A. L., Smith C. L., Raetz C. R., Kleppe K. Identity of the 17-kilodalton protein, a DNA-binding protein from Escherichia coli, and the firA gene product. J Bacteriol. 1988 Dec;170(12):5916–5918. doi: 10.1128/jb.170.12.5916-5918.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artz S. W., Broach J. R. Histidine regulation in Salmonella typhimurium: an activator attenuator model of gene regulation. Proc Natl Acad Sci U S A. 1975 Sep;72(9):3453–3457. doi: 10.1073/pnas.72.9.3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendiak D. S., Friesen J. D. Organization of genes in the four minute region of the Escherichia coli chromosome: evidence that rpsB and tsf are co-transcribed. Mol Gen Genet. 1981;181(3):356–362. doi: 10.1007/BF00425611. [DOI] [PubMed] [Google Scholar]
- Burgess R. R., Jendrisak J. J. A procedure for the rapid, large-scall purification of Escherichia coli DNA-dependent RNA polymerase involving Polymin P precipitation and DNA-cellulose chromatography. Biochemistry. 1975 Oct 21;14(21):4634–4638. doi: 10.1021/bi00692a011. [DOI] [PubMed] [Google Scholar]
- Carty M., Menzel R. Inhibition of DNA gyrase activity in an in vitro transcription-translation system stimulates gyrA expression in a DNA concentration dependent manner. Evidence for the involvement of factors which may be titrated. J Mol Biol. 1990 Jul 20;214(2):397–406. doi: 10.1016/0022-2836(90)90189-S. [DOI] [PubMed] [Google Scholar]
- Chamberlin M. J., Nierman W. C., Wiggs J., Neff N. A quantitative assay for bacterial RNA polymerases. J Biol Chem. 1979 Oct 25;254(20):10061–10069. [PubMed] [Google Scholar]
- Coleman J., Raetz C. R. First committed step of lipid A biosynthesis in Escherichia coli: sequence of the lpxA gene. J Bacteriol. 1988 Mar;170(3):1268–1274. doi: 10.1128/jb.170.3.1268-1274.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott T., Roth J. R. Characterization of Tn10d-Cam: a transposition-defective Tn10 specifying chloramphenicol resistance. Mol Gen Genet. 1988 Aug;213(2-3):332–338. doi: 10.1007/BF00339599. [DOI] [PubMed] [Google Scholar]
- Gonzalez N., Wiggs J., Chamberlin M. J. A simple procedure for resolution of Escherichia coli RNA polymerase holoenzyme from core polymerase. Arch Biochem Biophys. 1977 Aug;182(2):404–408. doi: 10.1016/0003-9861(77)90521-5. [DOI] [PubMed] [Google Scholar]
- Graña D., Gardella T., Susskind M. M. The effects of mutations in the ant promoter of phage P22 depend on context. Genetics. 1988 Oct;120(2):319–327. doi: 10.1093/genetics/120.2.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guterman S. K., Howitt C. L. Rifampicin supersensitivity of rho strains of E. coli, and suppression by sur mutation. Mol Gen Genet. 1979 Jan 16;169(1):27–34. doi: 10.1007/BF00267541. [DOI] [PubMed] [Google Scholar]
- Heil A., Zillig W. Reconstitution of bacterial DNA-dependent RNA-polymerase from isolated subunits as a tool for the elucidation of the role of the subunits in transcription. FEBS Lett. 1970 Dec;11(3):165–168. doi: 10.1016/0014-5793(70)80519-1. [DOI] [PubMed] [Google Scholar]
- Hirvas L., Coleman J., Koski P., Vaara M. Bacterial 'histone-like protein I' (HLP-I) is an outer membrane constituent? FEBS Lett. 1990 Mar 12;262(1):123–126. doi: 10.1016/0014-5793(90)80169-j. [DOI] [PubMed] [Google Scholar]
- Holck A., Kleppe K. Cloning and sequencing of the gene for the DNA-binding 17K protein of Escherichia coli. Gene. 1988 Jul 15;67(1):117–124. doi: 10.1016/0378-1119(88)90014-5. [DOI] [PubMed] [Google Scholar]
- Iwakura Y., Ishihama A., Yura T. RNA polymerase mutants of Escherichia coli. Streptolydigin resistance and its relation to rifampicin resistance. Mol Gen Genet. 1973 Mar 1;121(2):181–196. doi: 10.1007/BF00277531. [DOI] [PubMed] [Google Scholar]
- Jovanovich S. B., Lesley S. A., Burgess R. R. In vitro use of monoclonal antibodies in Escherichia coli S-30 extracts to determine the RNA polymerase sigma subunit required by a promoter. J Biol Chem. 1989 Mar 5;264(7):3794–3798. [PubMed] [Google Scholar]
- Koski P., Rhen M., Kantele J., Vaara M. Isolation, cloning, and primary structure of a cationic 16-kDa outer membrane protein of Salmonella typhimurium. J Biol Chem. 1989 Nov 15;264(32):18973–18980. [PubMed] [Google Scholar]
- Lathe R., Buc H., Lecocq J. P., Bautz E. K. Prokaryotic histone-like protein interacting with RNA polymerase. Proc Natl Acad Sci U S A. 1980 Jun;77(6):3548–3552. doi: 10.1073/pnas.77.6.3548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lathe R. Fine-structure mapping of the firA gene, a locus involved in the phenotypic expression of rifampin resistance in Escherichia. J Bacteriol. 1977 Sep;131(3):1033–1036. doi: 10.1128/jb.131.3.1033-1036.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lathe R., Lecocq J. P. The firA gene, a locus involved in the expression of rifampicin resistance in Escherichia coli. I. Characterisation of lambdafirA transducing phages constructed in vitro. Mol Gen Genet. 1977 Jul 7;154(1):43–51. doi: 10.1007/BF00265575. [DOI] [PubMed] [Google Scholar]
- Lathe R., Lecocq J. P. The firA gene, a locus involved in the expression of rifampicin resistance in Escherichia coli. II. Characterisation of bacterial proteins coded by lambdafirA transducing phages. Mol Gen Genet. 1977 Jul 7;154(1):53–60. doi: 10.1007/BF00265576. [DOI] [PubMed] [Google Scholar]
- Mulligan M. E., Hawley D. K., Entriken R., McClure W. R. Escherichia coli promoter sequences predict in vitro RNA polymerase selectivity. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 2):789–800. doi: 10.1093/nar/12.1part2.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson D., Weinstein M., Marshall S., Gillespie D. A new RNA synthesis mutant of E. coli. Biochem Genet. 1971 Dec;5(6):563–578. doi: 10.1007/BF00485674. [DOI] [PubMed] [Google Scholar]
- Rabussay D., Zillig W. A rifampicin resistent rna-polymerase from E. coli altered in the beta-subunit. FEBS Lett. 1969 Oct 21;5(2):104–106. doi: 10.1016/0014-5793(69)80305-4. [DOI] [PubMed] [Google Scholar]
- Thome B. M., Hoffschulte H. K., Schiltz E., Müller M. A protein with sequence identity to Skp (FirA) supports protein translocation into plasma membrane vesicles of Escherichia coli. FEBS Lett. 1990 Aug 20;269(1):113–116. doi: 10.1016/0014-5793(90)81132-8. [DOI] [PubMed] [Google Scholar]
- Watson R. J., Weis J. H., Salstrom J. S., Enquist L. W. Bacterial synthesis of herpes simplex virus types 1 and 2 glycoprotein D antigens. J Invest Dermatol. 1984 Jul;83(1 Suppl):102s–111s. doi: 10.1111/1523-1747.ep12281828. [DOI] [PubMed] [Google Scholar]
- Wiggs J. L., Bush J. W., Chamberlin M. J. Utilization of promoter and terminator sites on bacteriophage T7 DNA by RNA polymerases from a variety of bacterial orders. Cell. 1979 Jan;16(1):97–109. doi: 10.1016/0092-8674(79)90191-0. [DOI] [PubMed] [Google Scholar]
- Zagursky R., Baumeister K. Construction and use of pBR322 plasmids that yield single-stranded DNA for sequencing. Methods Enzymol. 1987;155:139–155. doi: 10.1016/0076-6879(87)55013-3. [DOI] [PubMed] [Google Scholar]