Abstract
Peroxisome proliferator-activated receptors (PPARs) α and γ are key regulators of lipid homeostasis and are activated by a structurally diverse group of compounds including fatty acids, eicosanoids, and hypolipidemic drugs such as fibrates and thiazolidinediones. While thiazolidinediones and 15-deoxy-Δ12,14-prostaglandin J2 have been shown to bind to PPARγ, it has remained unclear whether other activators mediate their effects through direct interactions with the PPARs or via indirect mechanisms. Here, we describe a novel fibrate, designated GW2331, that is a high-affinity ligand for both PPARα and PPARγ. Using GW2331 as a radioligand in competition binding assays, we show that certain mono- and polyunsaturated fatty acids bind directly to PPARα and PPARγ at physiological concentrations, and that the eicosanoids 8(S)-hydroxyeicosatetraenoic acid and 15-deoxy-Δ12,14-prostaglandin J2 can function as subtype-selective ligands for PPARα and PPARγ, respectively. These data provide evidence that PPARs serve as physiological sensors of lipid levels and suggest a molecular mechanism whereby dietary fatty acids can modulate lipid homeostasis.
Keywords: nuclear receptor, ligand, transcription, fibrate
While fatty acids (FAs) are essential biological components, elevated concentrations of circulating FAs are linked to a variety of disease states including obesity, atherosclerosis, and non-insulin-dependent diabetes mellitus (NIDDM) (1). Thus, physiologic levels of FAs levels must be maintained within narrow limits. Recent evidence suggests that FAs can rapidly modulate the transcription of genes involved in their own metabolism (2–4). Insight into the mechanism underlying some of these effects of FAs on gene expression was provided by the identification of several closely related orphan nuclear receptors, termed peroxisome proliferator-activated receptors (PPARs), that are activated by micromolar concentrations of a variety of FAs and FA analogues, including the hypolipidemic fibrate class of drugs (5–8). Three mammalian PPARs, designated PPARα, γ, and δ (9, 10), and three Xenopus laevis PPARs, designated xPPARα, -β, and -γ (11), have been isolated. While the PPARα and PPARγ subtypes are highly conserved across species, mammalian PPARδ and xPPARβ diverge significantly in sequence. Thus, it remains unclear whether these two subtypes are functional homologues. PPARs regulate the expression of target genes by binding to DNA sequence elements, termed PPAR response elements (PPREs). PPREs have been identified in the regulatory regions of a variety of genes that are involved in lipid metabolism and energy balance (9).
PPARα and PPARγ appear to play key roles in the catabolism and storage of fatty acids, respectively. Treatment of rodents with compounds that activate PPARα, including FAs, results in the proliferation of peroxisomes and the coordinate induction of hepatic genes involved in the β-oxidation of FAs in peroxisomes, mitochondria, and other cellular compartments (9, 12). Mice that lack functional PPARα do not respond to these agents and accumulate lipid droplets in their livers (13). Whereas PPARα functions in lipid catabolism and homeostasis in the liver, the PPARγ subtype appears to play a primary role in the storage of lipids in adipose tissue. PPARγ is abundantly expressed in adipocytes and plays a pivotal role in adipocyte differentiation in vitro (14–16).
We and others recently identified a class of antidiabetic drugs, the thiazolidinediones, as high-affinity PPARγ ligands (17, 18). The availability of radiolabeled thiazolidinediones provided the tool necessary for the subsequent discovery of the prostanoid 15-deoxy-Δ12,14-prostaglandin J2 (15d-J2) as a naturally occurring PPARγ ligand (18, 19). While a remarkable diversity of eicosanoids, FAs, and peroxisome proliferators have been reported to activate the PPARα subtype, the identification of bona fide PPARα ligands has been hindered by the lack of a comparable, high-affinity PPARα radioligand. In this report, we describe a novel fibrate that functions as a high-affinity ligand for both PPARα and PPARγ. Using this compound as a radioligand in competition binding assays, we demonstrate that specific FAs and eicosanoids can interact directly with both PPARα and PPARγ.
MATERIALS AND METHODS
Chemicals.
2-(4-[2-(3-[2,4-Difluorophenyl]-1-heptylureido) ethyl]phenoxy)-2-methylbutyric acid (GW2331) was synthesized via standard chemical techniques. [3H]GW2331 was synthesized by palladium catalyzed reduction of 2-(4-[2-(3-[2,4-difluorophenyl]-1-hept-2-enylureido)ethyl]phenoxy)-2-methylbutyric acid with tritium gas. The specific activity of the resulting radioligand was 82 Ci/mmol (1 Ci = 37 GBq). 15d-J2, 8(S)-hydroxyeicosatetraenoic acid (HETE), and 8(R)-HETE were purchased from Cayman Chemicals (Ann Arbor, MI). Wy 14,643 was purchased from Biomol (Plymouth Meeting, PA). All FAs were purchased from Sigma.
Cotransfection Assays.
To generate the pSG5-GAL4-xPPARα, pSG5-GAL4-xPPARβ, and pSG5-GAL4-xPPARγ chimeric receptor expression plasmids, cDNAs encoding the hinge and ligand binding domains of xPPARα (amino acids 174–474), xPPARβ (amino acids 96–396), and xPPARγ (amino acids 178–477) (11) were amplified by PCR and subcloned into the KpnI and BamHI sites of the pSG5-GAL4 expression plasmid (17). The clones were verified by sequence analysis. The pSG5-mouse (m) PPARα, pSG5-mPPARγ, pSG5-mPPARδ, pSG5-human (h) PPARα, pSG5-hPPARγ, and pSG5-hPPARδ chimeric receptor expression plasmids and the (UAS)5-tk-CAT reporter plasmid have been described (17, 19). Transient cotransfection assays using these plasmids were performed as described (17, 19).
Ligand Binding Assays.
To generate bacterial expression plasmids for the ligand binding domains of the PPAR subtypes, cDNAs encoding the hinge and ligand binding domains of hPPARα (amino acids 167–468) (20), mPPARα (amino acids 167–468) (5), xPPARα (amino acids 174–474) (11), and xPPARγ (amino acids 178–477) (11) were removed from the corresponding pSG-GAL4-PPAR chimeric receptor expression plasmids by cutting with KpnI and BamHI and inserted in frame into a pGEX-2T plasmid (Pharmacia) that had been modified to include KpnI and BamHI sites in the polylinker region. GST-PPAR fusion proteins or glutathione S-transferase (GST) alone as a control were expressed in BL21(DE3)plysS cells and extracts prepared by freeze-thawing the cells in bacterial lysis buffer (10 mM Tris, pH 8.0/250 mM KCl/1 mM DTT/1% Triton X-100) followed by centrifugation at 40,000 × g for 30 min. Glycerol was added to the bacterial extracts to a final concentration of 10%. Bacterial extracts were dialyzed extensively against bacterial lysis buffer containing 10% glycerol to remove glutathione that might interfere with the stability of the various FAs and eicosanoids in the competition binding assays. For saturation binding analysis or competition binding assays, bacterial extracts (≈50 μg protein) containing either GST-xPPARα or GST-xPPARγ were incubated at 4°C for 2–3 hr in buffer containing 10 mM Tris (pH 8.0), 50 mM KCl, and 10 mM DTT with [3H]GW2331 in the presence or absence of unlabeled GW2331 or the various FAs or eicosanoids. Bound radioactivity was separated from free radioactivity by elution through 1 ml Sephadex G-25 protein desalting columns (Boehringer Mannheim). Bound radioactivity eluted in the column void volume and was quantitated by liquid scintillation counting.
RESULTS
A Novel Fibrate Is a Potent PPARα and PPARγ Activator.
Drugs of the fibrate class are used clinically to reduce serum triglyceride and cholesterol levels and have been shown to activate the PPARs (11, 21–24). Recently, a novel class of trisubstituted ureido fibrate analogues was synthesized which displayed increased potency in rodent models of hyperlipidemia (D. Winegar, and S.S.S., unpublished data). We tested one of these fibrates, GW2331 (Fig. 1), for activity on the three human, mouse, and Xenopus PPAR subtypes in a transient transfection assay. To minimize background caused by endogenous PPARs present in CV-1 cells, an established chimera system was used in which the ligand binding domains of the PPARs were fused to the DNA binding domain of the yeast transcription factor GAL4 (17). Expression vectors encoding each of the chimeric receptors were transfected into CV-1 cells together with a reporter plasmid containing five copies of the GAL4 DNA binding site driving expression of the reporter chloramphenicol acetyltransferase (CAT). Interestingly, GW2331 was a potent activator of both the PPARα and PPARγ subtypes. Dose-response analysis revealed that GW2331 activated human, mouse, and Xenopus PPARα with EC50 values of 50 nM, 10 nM, and 60 nM, respectively (Table 1). GW2331 activated PPARγ from the different species with EC50 values that ranged from 200 nM to 360 nM (Table 1). In terms of efficacy, GW2331 activated PPARα and PPARγ to a degree comparable to that obtained with saturating concentrations of the efficacious activators Wy 14,643 and BRL 49653, respectively (data not shown). While GW2331 was only a weak activator of mouse and human PPARδ at doses up to 1 μM, GW2331 was a potent activator of xPPARβ (Table 1). Thus, GW2331 represents a novel fibrate that functions as a dual activator of PPARα and PPARγ from multiple species as well as xPPARβ.
Figure 1.
Structure of the fibrate GW2331.
Table 1.
EC50 values for activation of Xenopus, mouse, and human PPARs by the fibrate GW2331
| Species | EC50 values, nM
|
||
|---|---|---|---|
| PPARα | PPARγ | PPARβ/δ | |
| Human | 50 | 300 | >1,000 |
| Mouse | 10 | 200 | >1,000 |
| Xenopus | 60 | 360 | 40 |
EC50 values were determined using the GAL4-PPAR chimeras. CV-1 cells were transfected with the various GAL4-PPAR expression plasmids and the cells were treated with GW2331 at concentrations ranging from 1 × 10−10 M to 1 × 10−5 M in 1/3 molar log steps. All data points were tested in triplicate. The overall levels of activation of PPARα and PPARγ were comparable to those obtained with saturating concentrations of Wy 14,643 and BRL 49653, respectively. Similar EC50 values were obtained in two independent experiments.
GW2331 Is a High-Affinity PPARα and PPARγ Ligand.
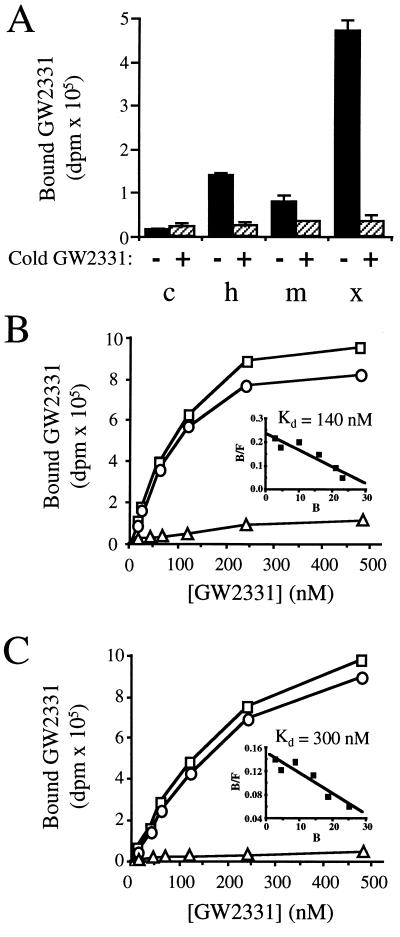
We next sought to determine whether GW2331 interacted directly with PPARα. The putative ligand binding domains of human, mouse, and Xenopus PPARα were overexpressed in Escherichia coli as fusion proteins with GST. Bacterial extracts containing GST-hPPARα, GST-mPPARα, or GST-xPPARα or control extracts containing GST alone were tested for binding to [3H]GW2331. Specific binding of [3H]GW2331 was detected in extracts containing each of the three fusion proteins (Fig. 2A). Significantly more binding activity was detected in extracts containing GST-xPPARα than those containing either GST-hPPARα or GST-mPPARα (Fig. 2A). Consistent with this finding, Western blot analysis revealed markedly higher levels of GST-xPPARα than either GST-hPPARα or GST-mPPARα in the soluble fraction of bacterial lysates (data not shown). In control experiments, no specific binding of [3H]GW2331 was detected in bacterial extracts containing GST alone (Fig. 2A).
Figure 2.
[3H]GW2331 binds to PPARα and PPARγ. (A) Control bacterial extracts (c) or bacterial extracts containing GST-hPPARα (h), GST-mPPARα (m), or GST-xPPARα (x) were incubated with 60 nM [3H]GW2331 in either the absence or presence of 10 μM unlabeled GW2331 as indicated. The amount of GST included in the control experiments was comparable or exceeded the amount of GST-PPAR fusion proteins used in binding experiments as judged by SDS/PAGE analysis and Coomassie blue staining of bacterial extracts (data not shown). Data shown represent the mean of assays performed in triplicate ± SD. (B) Bacterial extracts containing GST-xPPARα were incubated with increasing concentrations of [3H]GW2331 in the absence (total binding; □) or presence (nonspecific binding; ▵) of 10 μM unlabeled GW2331. Specific binding is indicated (○). (Inset) Specific binding data were transformed by Scatchard analysis. (C) Bacterial extracts containing GST-xPPARγ were incubated with increasing concentrations of [3H]GW2331 in the absence (total binding; □) or presence (nonspecific binding; ▵) of 10 μM unlabeled GW2331. Specific binding is indicated (○). (Inset) Specific binding data were transformed by Scatchard analysis.
Based upon the higher specific binding activity obtained using bacterial extracts containing the Xenopus receptor relative to the human and murine homologues, the GST-xPPARα protein was used to further characterize the binding properties of GW2331. Saturation binding analyses revealed that [3H]GW2331 bound in a saturable manner to GST-xPPARα with a Kd value of 140 nM (Fig. 2B). These data are in good agreement with the EC50 value for xPPARα activation obtained using the transient transfection assay (Table 1) and demonstrate that a molecule of the fibrate class can function as a high-affinity ligand for PPARα. In parallel studies, we found that GW2331 also interacted with xPPARγ. [3H]GW2331 bound specifically to a GST-xPPARγ fusion protein with a Kd value of 300 nM (Fig. 2C). [3H]GW2331 also bound specifically to the analogous GST-mPPARγ fusion protein (data not shown). Thus, it appears that GW2331 represents the first molecule to be identified that binds with high-affinity to both the PPARα and PPARγ subtypes.
FAs Bind to PPARα and PPARγ.
The availability of [3H]GW2331 as a high-affinity ligand for both PPARα and PPARγ provided us with the means to examine whether activators of these receptors are in fact ligands. We initially examined a variety of FAs for their ability to activate xPPARα and xPPARγ in the transfection assay and, in parallel, their ability to compete with [3H]GW2331 for binding to GST-xPPARα and GST-xPPARγ. Because the total concentration of nonesterified FAs in adult, human serum can exceed 1 mM and the more prevalent of the individual FAs account for 20–40% of this total (25), we assayed the individual FAs at a concentration of 30 μM in the competition binding assay and 100 μM in the transfection assay.
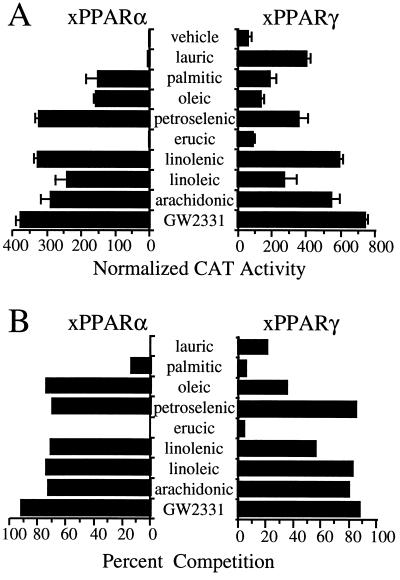
Transfection analysis and competition binding assays were performed with the saturated FAs lauric (C12:0) and palmitic acid (C16:0); the monounsaturated FAs oleic (C18:1), petroselenic acid (C18:1), and erucic acid (C22:1); and, the polyunsaturated (PU) FAs linolenic (C18:3), linoleic (C18:2), and arachidonic (C20:4) acids. With the exceptions of lauric and erucic acid, each of these FAs was a strong activator of the GAL4-xPPARα chimera in the transactivation assay when assayed at a concentration of 100 μM (Fig. 3A). The more efficacious of these FAs activated xPPARα to levels comparable to those obtained with GW2331 (Fig. 3A). As we have previously observed with the mammalian PPAR subtypes (S.A.K. and J.M.L., unpublished observation), the basal activity of xPPARγ was markedly higher (≈30-fold) than that of xPPARα (Fig. 3A). However, xPPARγ activity was augmented in the presence of 100 μM lauric acid, petroselenic acid, and each of the PUFAs (Fig. 3A). Palmitic and oleic acid were weak activators, while erucic acid showed virtually no activity on xPPARγ (Fig. 3A). Similar results were obtained in the presence of 100 μM of these FAs when mPPARα or mPPARγ were substituted for the Xenopus homologues in the transient transfection experiments (data not shown). These data are consistent with previous studies demonstrating that mammalian and Xenopus PPARs are activated by micromolar concentrations of a structurally diverse group of FAs (6–8, 23, 26).
Figure 3.
FAs are ligands for xPPARα and xPPARγ. (A) CV-1 cells were cotransfected with expression plasmids encoding either GAL4-xPPARα or GAL4-xPPARγ and the reporter plasmid UAS5-tk-CAT. Cells were treated with vehicle (0.1% dimethyl sulfoxide; DMSO) alone, 1 × 10−4 M of the various FAs, or 1 × 10−6 M GW2331. Cell extracts were subsequently assayed for CAT activity. Data points represent the mean of assays performed in triplicate. Similar results were obtained in two independent experiments. (B) Competition binding assays were performed using bacterial extracts containing either GST-xPPARα or GST-xPPARγ and either 30 nM (xPPARα) or 60 nM (xPPARγ) [3H]GW2331 in the presence of 3 × 10−5 M of the indicated FAs or unlabeled GW2331. Data are presented as percent competition relative to addition of vehicle (1% DMSO) alone and represent the mean of assays performed in triplicate. Similar results were obtained in two independent experiments.
In competition binding assays, we found that xPPARα and xPPARγ were also remarkably promiscuous in terms of their direct interactions with FAs. All three PUFAs competed efficiently with [3H]GW2331 for binding to both xPPARα and xPPARγ (Fig. 3B). The monounsaturated FA petroselenic acid also competed efficiently with [3H]GW2331 for binding to both Xenopus receptors, whereas oleic acid, which differs from petroselenic acid only in the position of the single double bond, competed significantly better for binding to xPPARα than xPPARγ (Fig. 3B). In agreement with the transfection data, little or no competition was seen with erucic acid on either of the two PPARs (Fig. 3B). Surprisingly, however, lauric acid, which activates xPPARγ, and palmitic acid, which activates both PPARs, competed only weakly with [3H]GW2331 in the competition binding assay (Fig. 3B). We speculate that these FAs may either serve as precursors to more active compounds or, alternatively, interfere with cellular metabolism, and result in the accumulation of endogenous PPAR ligands. We conclude from these data that xPPARα and xPPARγ are bona fide receptors for a structurally diverse set of FAs, including mono- and PUFAs, but that not all activators mediate their effects through direct interactions with the PPARs.
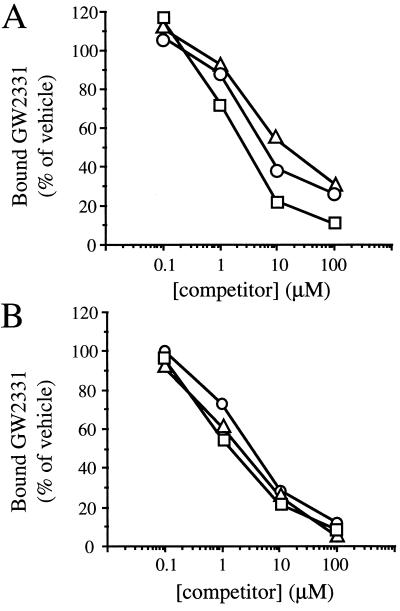
Based on their efficacies in the competition binding assay and relative abundance in human serum (25), we selected the PUFAs linolenic, linoleic, and arachidonic acid for more thorough dose response analysis. These three FAs competed with [3H]GW2331 for binding to GST-xPPARα and GST-xPPARγ with IC50 values in the 2-to 20-μM range (Fig. 4 A and B). These values are within the range of concentrations of the nonesterified FAs found in human serum (25) suggesting that these dietary FAs serve as PPAR ligands in vivo.
Figure 4.
PUFAs are ligands for xPPARα and xPPARγ. Competition binding assays were performed with (A) GST-xPPARα and 30 nM [3H]GW2331 or (B) GST-xPPARγ and 60 nM [3H]GW2331 in the presence of increasing concentrations of linolenic acid (▵), linoleic acid (○), or arachidonic acid (□). Data represent the mean of duplicate points and were normalized to reactions performed with vehicle (1% DMSO) alone.
Eicosanoids Are Subtype Selective PPAR Ligands.
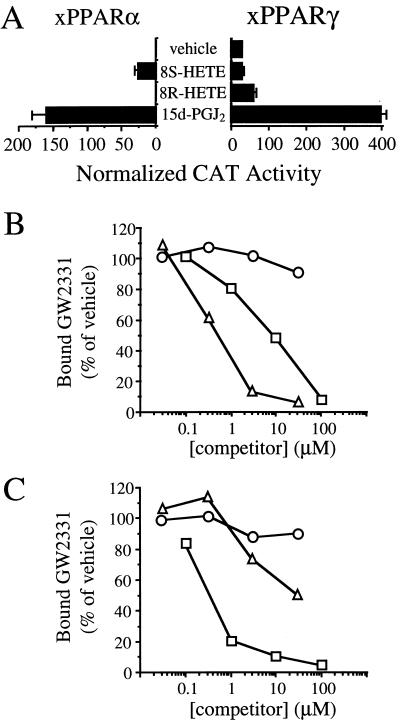
The PPARs have recently been found to be activated by various eicosanoid and eicosanoid-like molecules (7, 18, 19, 26–30). We chose two naturally occurring eicosanoids, 15d-J2 and 8-HETE, for evaluation in the transactivation and competition binding assays. 15d-J2 has been shown to bind and activate PPARγ and, at higher concentrations, to activate PPARα (18, 19). 8-HETE is an 8-lipoxygenase product that is produced in skin in response to treatment with phorbol esters (31–33) and was recently shown to be a selective activator of mPPARα (27). Interestingly, the 8(S)-HETE stereoisomer was found to be significantly more active than its 8(R) enantiomer in activating mPPARα in transfection assays (27). In agreement with these previous studies, we found that the (S) enantiomer of 8-HETE was a selective xPPARα activator and that 15d-J2 activated both xPPARα and xPPARγ when these eicosanoids were tested in transfection assays at a concentration of 1 × 10−5 M (Fig. 5A).
Figure 5.
Eicosanoids are selective xPPARα and xPPARγ ligands. (A) CV-1 cells were cotransfected with expression plasmids encoding either GAL4-xPPARα or GAL4-xPPARγ and the reporter plasmid UAS5-tk-CAT. Cells were treated with vehicle (0.1% DMSO) alone or 1 × 10−5 M of the eicosanoids 8(S)-HETE, 8(R)-HETE, or 15d-J2. Cell extracts were subsequently assayed for CAT activity. (B and C) Competition binding assays were performed with (B) GST-xPPARα and 30 nM [3H]GW2331 or (C) GST-xPPARγ and 60 nM [3H]GW2331 in the presence of increasing concentrations of 8(S)-HETE (▵), 8(R)-HETE (○), or 15d-J2 (□). Data represent the mean of duplicate points and were normalized to reactions performed with vehicle (1% DMSO) alone.
We next tested these eicosanoids in the competition binding assay. In agreement with results obtained with mammalian PPARγ, 15d-J2 interacted with GST-xPPARγ with an IC50 value of roughly 500 nM (Fig. 5C). 15d-J2 was one to two orders of magnitude more potent in its interaction with GST-xPPARγ than with GST-xPPARα (Fig. 5 B and C). Whereas 15d-J2 interacted preferentially with xPPARγ, 8(S)-HETE displayed a strong preference for interacting with xPPARα. 8(S)-HETE interacted with GST-xPPARα with an IC50 value of ≈500 nM (Fig. 5B). By contrast, 8(S)-HETE interacted only weakly with GST-xPPARγ (Fig. 5C). The (R) enantiomer of 8-HETE failed to compete efficiently with [3H]GW2331 for binding to either GST-xPPARα or GST-xPPARγ (Fig. 5 B and C). We conclude that the naturally occurring eicosanoids 8(S)-HETE and 15d-J2 are both more potent and more subtype-selective in their interactions with the PPARs than are their FA precursors.
DISCUSSION
While many natural and synthetic compounds have been shown to activate the PPARs, their biophysical properties and lack of potency has made them largely unsuitable for use in ligand binding assays. Thus, in the case of PPARα, it has remained unclear whether these activators mediate their effects through direct interactions with this orphan receptor or via an indirect mechanism. We now report the identification of the fibrate GW2331 as a high-affinity PPARα and PPARγ ligand. Through the use of radiolabeled GW2331 in competition binding assays, we show that several FAs that are prevalent in vivo, including the monounsaturated FA oleic acid and the PUFAs linoleic acid, linolenic acid, and arachidonic acid can function as ligands for both PPARα and PPARγ at concentrations that are consistent with those found in human serum (25). These data provide strong evidence that FAs serve as endogenous ligands for PPARα and PPARγ. Moreover, our findings suggest the existence of a molecular feedback mechanism whereby increases in the circulating levels of these FAs could result in enhanced catabolism and storage of FAs via activation of PPARα and PPARγ, respectively.
The finding that various FAs can function as PPARα and PPARγ ligands at micromolar concentrations does not exclude the possibility of the existence of either higher affinity or more subtype-selective PPAR ligands. Indeed, we have shown that the PUFA metabolites 8(S)-HETE and 15d-J2 are potent, selective activators of PPARα and PPARγ, respectively. Thus, the regulated conversion of PUFAs to eicosanoids through either the lipoxygenase or cyclooxygenase pathways may provide a mechanism for the differential regulation of PPARα and PPARγ and their respective target genes. Unlike FAs, which are quite stable under standard extraction conditions, eicosanoids such as 8(S)-HETE and 15d-J2 are extremely labile. Thus, it remains unclear whether these eicosanoids are present in vivo at sufficient concentrations to serve as PPAR ligands.
As receptors for various FAs and eicosanoids, the PPARs appear to be much more promiscuous in their interactions with ligands than other members of the nuclear receptor family. Our results suggest that the degree of activation of the PPARs in vivo may not be determined simply through interactions with a single, high-affinity ligand, but may instead be a function of the sum concentration of a variety of FAs and FA metabolites that can interact with the receptors. While the effective concentrations of FAs in vivo are difficult to ascertain due to their interactions with intracellular and extracellular binding proteins, the net concentration of nonesterified FAs in normal adult human serum is in the range necessary to activate the PPARs (25). This idea is supported by a recent study in which FAs, including arachidonic, linoleic, and oleic acids, were isolated via gas chromatography-mass spectrometry as the components present in human serum that activate PPARα (34).
We have shown that certain mono- and PUFAs can interact directly with PPARα and PPARγ. Interestingly, diets rich in these unsaturated FAs have been reported to lower serum cholesterol and triglyceride levels and elevate high density lipoprotein cholesterol levels in animals and humans (1, 35, 36). There is also evidence that these FAs may provide protection against cardiovascular disease (1). Furthermore, increased insulin sensitivity was found to be associated with increased PUFA levels in skeletal muscle-phospholipids in humans (37), and PUFAs were found to prevent diet-induced insulin resistance in rodents (38). Insulin resistance is a central feature of NIDDM. The pharmacological actions of the synthetic PPAR ligands and activators support the idea that mono- and PUFAs may mediate some of their effects through the PPARs. Fibrates, which activate PPARα (5) and in some cases also activate PPARγ (11, 24), demonstrate a broad spectrum of effects on lipids in man including reductions in total plasma cholesterol and triglyceride levels (21, 22). The antidiabetic thiazolidinediones, which are selective PPARγ ligands, enhance the actions of insulin in peripheral tissues and also reduce serum lipid levels in rodents and humans suffering from NIDDM (39, 40). The parallels seen between the effects of mono- and PUFAs and the synthetic PPAR ligands suggest that PPARs may be the molecular targets for at least some of the beneficial effects of the dietary FAs.
In summary, we have demonstrated that a subset of eicosanoids and FAs, including essential FAs, can bind directly to PPARα and PPARγ. These data ascribe a novel regulatory function to FAs and certain of their cyclooxygenase and lipoxygenase metabolites. Moreover, as PPARα and PPARγ appear to play central roles in the catabolism and storage of FAs, our data suggest a molecular mechanism whereby dietary lipids could affect overall energy balance and impact such metabolic diseases as obesity, atherosclerosis, and NIDDM.
Acknowledgments
We thank S. Prakash and Itzela Correa for purification and characterization of [3H]GW2331; Ken Batchelor and Bill Wilkison for discussions and support; Jim Chapman (University of South Carolina), Roy Hawke, and Deborah Winegar for communicating data prior to publication; and Barry Forman and Ron Evans for sharing results prior to publication.
ABBREVIATIONS
- CAT
chloramphenicol acetyl-transferase
- 15d-J2
15-deoxy-Δ12,14-prostaglandin J2: FA, fatty acid
- GST
glutathione S-transferase
- HETE
hydroxyeicosatetraenoic acid
- PPAR
peroxisome proliferator-activated receptor
- x
Xenopus laevis
- m
mouse
- h
human
- PU
polyunsaturated
- NIDDM
non-insulin-dependent diabetes mellitus
- GW2331
2-(4-[2-(3-[2,4-difluorophenyl]-1-heptylureido)ethyl]phenoxy)-2-methylbutyric acid
- DMSO
dimethyl sulfoxide
References
- 1.Horrobin D F. Prostaglandins Leukotrienes Essent Fatty Acids. 1995;53:385–396. doi: 10.1016/0952-3278(95)90101-9. [DOI] [PubMed] [Google Scholar]
- 2.Clarke S D, Jump D B. Lipids. 1996;37:S7–S11. doi: 10.1007/BF02637044. [DOI] [PubMed] [Google Scholar]
- 3.Hillgartner F B, Salati L M, Goodridge A G. Physiol Rev. 1995;75:47–76. doi: 10.1152/physrev.1995.75.1.47. [DOI] [PubMed] [Google Scholar]
- 4.Amri E-Z, Ailhaud G, Grimaldi P-A. J Lipid Res. 1994;35:930–937. [PubMed] [Google Scholar]
- 5.Issemann I, Green S. Nature (London) 1990;347:645–650. doi: 10.1038/347645a0. [DOI] [PubMed] [Google Scholar]
- 6.Göttlicher M, Widmark E, Li Q, Gustafsson J-A. Proc Natl Acad Sci USA. 1992;89:4653–4657. doi: 10.1073/pnas.89.10.4653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Keller H, Dreyer C, Medin J, Hahfoudi A, Ozato K, Wahli W. Proc Natl Acad Sci USA. 1993;90:2160–2164. doi: 10.1073/pnas.90.6.2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Krey G, Keller H, Mahfoudi A, Medin J, Ozato K, Dreyer C, Wahli W. J Steroid Biochem Mol Biol. 1993;47:65–73. doi: 10.1016/0960-0760(93)90058-5. [DOI] [PubMed] [Google Scholar]
- 9.Desvergne B, Wahli W. In: Inducible Gene Expression. Baeuerle P A, editor. Boston: Birkhauser; 1995. pp. 142–176. [Google Scholar]
- 10.Green S, Wahli W. Mol Cell Endocrinol. 1994;100:149–153. doi: 10.1016/0303-7207(94)90294-1. [DOI] [PubMed] [Google Scholar]
- 11.Dreyer C, Krey G, Keller H, Givel F, Helftenbein G, Wahli W. Cell. 1992;68:879–887. doi: 10.1016/0092-8674(92)90031-7. [DOI] [PubMed] [Google Scholar]
- 12.Reddy J K, Goel S K, Nemali M R, Carrino J J, Laffler T G, Reddy M K, Sperbeck S J, Osumi T, Hashimoto T, Lalwani N D, Rao M S. Proc Natl Acad Sci USA. 1986;83:1747–1751. doi: 10.1073/pnas.83.6.1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee S S-T, Pineau T, Drago J, Lee E J, Owens J W, Kroetz D L, Fernandez-Salguero P M, Westphal H, Gonzalez F J. Mol Cell Biol. 1995;15:3012–3022. doi: 10.1128/mcb.15.6.3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tontonoz P, Hu E, Graves R, Budavari A, Spiegelman B. Genes Dev. 1994;8:1224–1234. doi: 10.1101/gad.8.10.1224. [DOI] [PubMed] [Google Scholar]
- 15.Tontonoz P, Hu E, Spiegelman B M. Cell. 1994;79:1147–1156. doi: 10.1016/0092-8674(94)90006-x. [DOI] [PubMed] [Google Scholar]
- 16.Chawla A, Schwartz E J, Dimaculangan D D, Lazar M A. Endocrinology. 1994;135:798–800. doi: 10.1210/endo.135.2.8033830. [DOI] [PubMed] [Google Scholar]
- 17.Lehmann J M, Moore L B, Smith-Oliver T A, Wilkison W O, Willson T M, Kliewer S A. J Biol Chem. 1995;270:12953–12956. doi: 10.1074/jbc.270.22.12953. [DOI] [PubMed] [Google Scholar]
- 18.Forman B, Tontonoz P, Chen J, Brun R, Spiegelman B, Evans R. Cell. 1995;83:803–812. doi: 10.1016/0092-8674(95)90193-0. [DOI] [PubMed] [Google Scholar]
- 19.Kliewer S A, Lenhard J M, Willson T M, Patel I, Morris D C, Lehmann J M. Cell. 1995;83:813–819. doi: 10.1016/0092-8674(95)90194-9. [DOI] [PubMed] [Google Scholar]
- 20.Sher T, Yi H-F, McBride O W, Gonzalez F J. Biochemistry. 1993;32:5598–5604. doi: 10.1021/bi00072a015. [DOI] [PubMed] [Google Scholar]
- 21.Catapano A L. Pharmacol Rev. 1992;26:331–340. doi: 10.1016/1043-6618(92)90232-z. [DOI] [PubMed] [Google Scholar]
- 22.Sirtori C R, Franceschini G. Pharmacol Ther. 1988;37:167–191. doi: 10.1016/0163-7258(88)90024-1. [DOI] [PubMed] [Google Scholar]
- 23.Issemann I, Prince R A, Tugwood J D, Green S. J Mol Endocrinol. 1993;11:37–47. doi: 10.1677/jme.0.0110037. [DOI] [PubMed] [Google Scholar]
- 24.Zhu Y, Alavares K, Huang Q, Rao M S, Reddy J K. J Biol Chem. 1993;268:26817–26820. [PubMed] [Google Scholar]
- 25.Jüngling E, Kammermeier H. Anal Biochem. 1988;171:150–157. doi: 10.1016/0003-2697(88)90136-4. [DOI] [PubMed] [Google Scholar]
- 26.Kliewer S A, Forman B M, Blumberg B, Ong E S, Borgmeyer U, Mangelsdorf D J, Umesono K, Evans R M. Proc Natl Acad Sci USA. 1994;91:7355–7359. doi: 10.1073/pnas.91.15.7355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu K, Bayona W, Kallen C B, Harding H P, Ravera C P, McMahon G, Brown M, Lazar M A. J Biol Chem. 1995;270:23975–23983. doi: 10.1074/jbc.270.41.23975. [DOI] [PubMed] [Google Scholar]
- 28.Hertz R, Berman I, Keppler D, Bar-Tana J. Eur J Biochem. 1996;235:242–247. doi: 10.1111/j.1432-1033.1996.00242.x. [DOI] [PubMed] [Google Scholar]
- 29.Brun R P, Tontonoz P, Forman B M, Ellis R, Chen J, Evans R M, Spiegelman B M. Genes Dev. 1996;10:974–984. doi: 10.1101/gad.10.8.974. [DOI] [PubMed] [Google Scholar]
- 30.Devchand P R, Keller H, Peters J M, Vazquez M, Gonzalez F J, Wahli W. Nature (London) 1996;384:39–43. doi: 10.1038/384039a0. [DOI] [PubMed] [Google Scholar]
- 31.Gschwendt M, Furstenberger G, Kittstein W, Besemfelder E, Hull W E, Hagedorn H, Opferkuch H J, Marks F. Carcinogenesis. 1986;7:449–455. doi: 10.1093/carcin/7.3.449. [DOI] [PubMed] [Google Scholar]
- 32.Furstenberger G, Hagedorn H, Jacobi T, Besemfelder E, Stephan M, Lehmann W D, Marks F. J Biol Chem. 1991;266:15738–15745. [PubMed] [Google Scholar]
- 33.Hughes M A, Brash A R. Biochim Biophys Acta. 1991;1081:347–354. doi: 10.1016/0005-2760(91)90292-p. [DOI] [PubMed] [Google Scholar]
- 34.Banner C D, Göttlicher M, Widmark E, Sjövall J, Rafter J J, Gustafsson J-A. J Lipid Res. 1993;34:1583–1591. [PubMed] [Google Scholar]
- 35.Grundy S, Denke M A. J Lipid Res. 1990;31:1149–1172. [PubMed] [Google Scholar]
- 36.Nestel P J. Annu Rev Nutr. 1990;10:149–167. doi: 10.1146/annurev.nu.10.070190.001053. [DOI] [PubMed] [Google Scholar]
- 37.Borkman M, Storlien L H, Pan D A, Jenkins A B, Chisholm D J, Campbell L V. N Engl J Med. 1993;328:238–244. doi: 10.1056/NEJM199301283280404. [DOI] [PubMed] [Google Scholar]
- 38.Storlien L H, Kraegen E W, Chisholm D J, Ford G L, Bruce D G, Pascoe W S. Science. 1987;237:885–888. doi: 10.1126/science.3303333. [DOI] [PubMed] [Google Scholar]
- 39.Saltiel A R, Olefsky J M. Diabetes. 1996;45:1661–1669. doi: 10.2337/diab.45.12.1661. [DOI] [PubMed] [Google Scholar]
- 40.Nolan J, Ludvik B, Beerdsen P, Joyce M, Olefsky J. N Engl J Med. 1994;331:1188–1193. doi: 10.1056/NEJM199411033311803. [DOI] [PubMed] [Google Scholar]