Summary
Insect pheromone binding proteins (PBPs) transport sex pheromones through the aqueous layer surrounding G-protein coupled receptors that initiate signaling events leading to mating. This PBP-receptor system strongly discriminates between ligands with subtle structural differences, but it has proved difficult to distinguish the degree of discrimination of the PBP from that of the G-protein coupled receptor. The three-dimensional structures of the PBP of Bombyx mori, the silkworm moth, both with and without its cognate ligand, bombykol [(E,Z)-10,12-hexadecadienol], have been determined by X-ray crystallography and NMR. In this paper, the structures of the same binding protein with bound iodohexadecane and bell pepper odorant, were determined at 1.9 Å and 2.0 Å, respectively. These structures illustrate the remarkable plasticity in the ligand binding site of the PBP but suggest the protein might still act as a filter during pheromone signal processing.
Introduction
Sex pheromone signaling in moths forms a paradigm for chemical communication: a female moth sends out a chemical message that, upon being encountered by an appropriate male, is processed to generate a correct physiological response within milliseconds. The sex pheromone of Bombyx mori, bombykol [(E,Z)-10,12-hexadecadienol, Fig. 1], is synthesized by the female and detected in the highly branched antennae of male moths. Pheromone is adsorbed into specialized male moth olfactory hairs, or sensilla, which cover the antennae. The pheromone diffuses through pore tubules into the aqueous sensillar lymph where it is bound to the pheromone binding protein and transported to the G-protein coupled receptor on the neural cell. Using this signaling system, the male follows the pheromone plume to its mate. Transporting the hydrophobic pheromone through the aqueous sensillar lymph could delay the rapid response needed to follow the pheromone plume. The lymph of pheromone-sensitive olfactory hairs in Bombyx mori antennae contains a high concentration of pheromone-binding proteins (PBPs), 10-20 mM or about 160 mg/mL (Vogt, 1987). PBPs are members of the encapsulins (Leal, 2003), a family of proteins that solubilize hydrophobic compounds in an aqueous environment. The subclass of pheromone binding proteins mediates the delivery of the sex pheromone to its receptor in the dendritic membrane.
Figure 1.
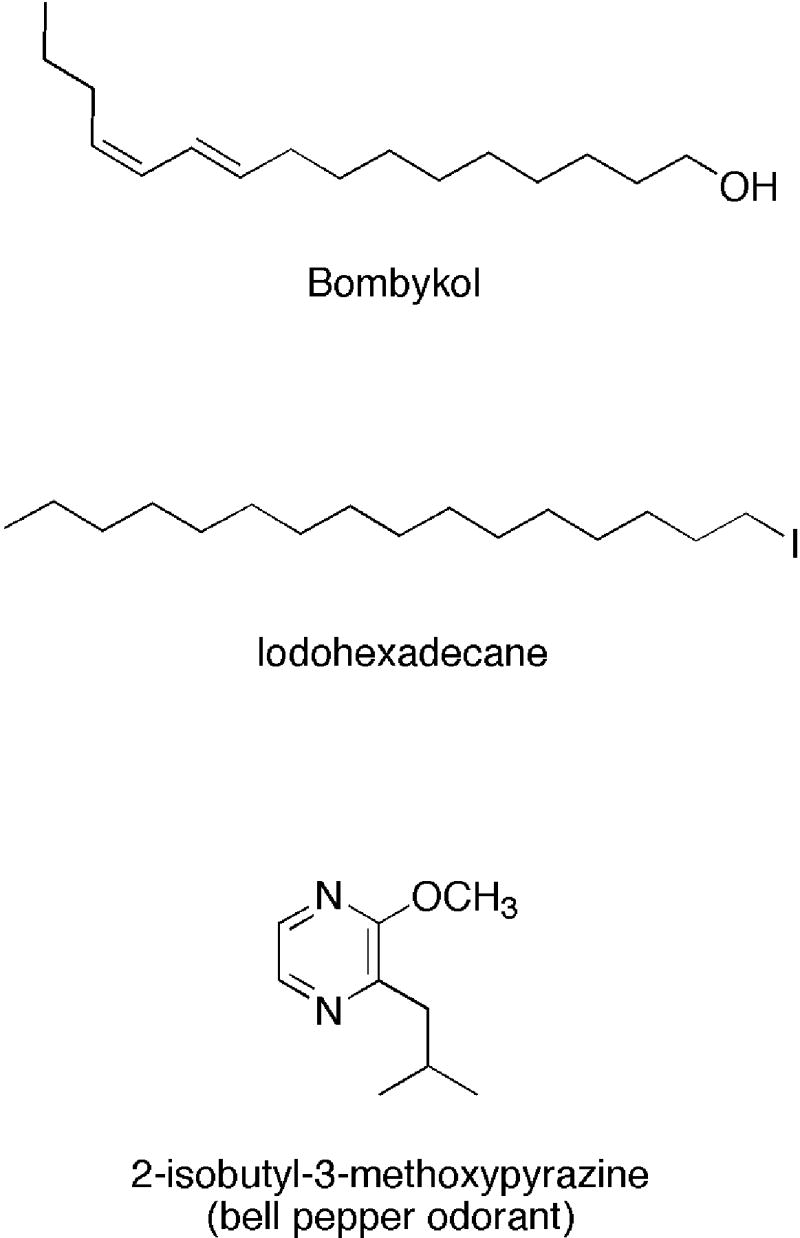
Ligands Used for Cocrystallization with BmorPBP
Pheromone reception in male moths is both highly sensitive – responses can be seen with only a few molecules – and selective – small structural changes typically lead to loss of activity by several orders of magnitude. This ligand selectivity is essential given the degree of similarity of moth sex pheromones, which are typically twelve- to eighteen-carbon partially unsaturated aliphatic chains (Ando, 2004). These molecules differ only by carbon chain length, placement of double bonds, and terminal functional group (aldehyde, alcohol, or acetate). Early studies, for example, indicated that simply switching either the cis or trans double bond in bombykol rendered the molecule ineffective as an attractant (Butenandt, 1963). It has been suggested that PBPs play a role in discriminating among potential molecular signals (Leal, 2003, 2005; Pelosi, 1994; Prestwich, 1997) and in speeding up the pheromone signaling process (Syed et al., 2006).
There is evidence both supporting and contradicting the idea that PBPs are involved in pheromone recognition. Analysis of the primary structure of PBPs from different moth species shows limited diversity among the proteins. The sequences are about 70% identical and 85% similar to each other, and sequence-based searching for amino acid residues that might be involved in specificity has yielded little information, although sequence and structural data suggest that serine residues interact with alcohol groups in 14-16 carbon chain pheromones, and that asparagine residues might specifically interface with acetate groups (Mohanty et al., 2004; Sandler et al., 2000). However, for most moth species the pheromone signal consists of several molecules and several PBPs. It has been demonstrated through binding studies that for Antheraea polyphemus and A. pernyi moths, each of which has three pheromone binding proteins and a three-component pheromone blend, that each PBP preferentially binds a specific component of the blend (Maida et al., 2003). All three constituents of the sex pheromone of the wild silkmoth, A. polyphemus, bound to the major PBP from this species, ApolPBP1, with apparent high affinity, but in competitive assays ApolPBP1 showed considerable preference for the major constituent of the sex pheromone.
We felt that structural studies with ligands very different from pheromones might offer some insight into PBP specificity and plasticity. To explore the specificity of B. mori PBP (BmorPBP), the protein was crystallized in complex with non-pheromone ligands (Fig. 1) and structures were determined by X-ray crystallography. To investigate limitations that would be imposed by stringent specificity, iodohexadecane was added to form the IHD-BmorPBP complex. Iodohexadecane has a chain length slightly longer than that of bombykol, lacks the conformational restraint provided by the two double bonds in the pheromone, replaces the alcohol functional group with an iodine atom, and has no detectable pheromonal activity. To explore a potential ligand with quite different geometry than that of a typical moth sex pheromone, bell pepper odorant (2-isobutyl-3-methoxypyrazine) was added to form BPO-BmorPBP complex. The bell pepper odorant molecule lacks the long chain character of bombykol but retains the hydrophobic nature of sex pheromones, seems to be a promiscuous binder of the closely-related odorant binding protein family, and has no reported pheromone role. Both crystal structures revealed electron density in the binding pocket of the protein that precisely fit the geometry of each added non-pheromone ligand. While both of these differently-shaped hydrophobic ligands are bound, the structures also provide experimental information that suggests the binding pocket of the protein is adapted for ligands with hook-shaped geometries, such as that of bombykol.
Results
Overall structures
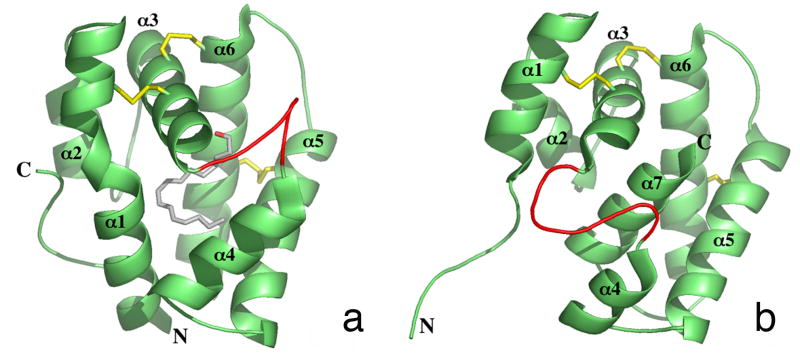
All structures were solved by molecular replacement using the crystal structure of B. mori PBP bound to bombykol at pH 8 [Fig. 2a (Sandler et al., 2000)]. Bombykol was removed from the model for molecular replacement. In the bombykol complex, the protein is comprised of six α-helices. Four of these six helices converge to form a hydrophobic binding pocket for bombykol, with three disulfide bonds stabilizing the structure of this small extracellular protein. A loop region between helices α3 and α4 is believed to provide entrance for the ligand by becoming destabilized upon protonation of one or all of three histidine residues at low pH (Sandler et al., 2000). The bound bombykol has a roughly planar, hook-shaped conformation within the binding pocket. The hydroxyl-group of bombykol forms a hydrogen bond with the sidechain of a Ser56, and one set of double bonds in bombykol is sandwiched between the Phe12 and Phe118 aromatic rings.
Figure 2. Crystal Structures of Bombykol-Bound and Unliganded BmorPBP.
X-ray crystal structures showing bombykol (A) or no ligand (B) in the binding pocket of BmorPBP (Protein Data Bank ID codes 1DQE and 2FJY, respectively). Secondary structures are depicted in cartoon representation. Bombykol is depicted in ball-and-stick format. Pictures were generated in PyMOL (http://www.pymol.org/).
The protein has also been shown to exist in an empty structure [Fig. 2b, (Lautenschlager et al., 2005)]. The bombykol-bound protein and empty protein differ structurally in three major ways: the bombykol-bound protein has a disordered C-terminus, while the empty protein has a disordered N-terminus; the disordered looping region between helices α3 and α4 is more extended in the empty protein; and the C-terminus of the empty protein forms a seventh α-helix that fills the binding pocket of the protein.
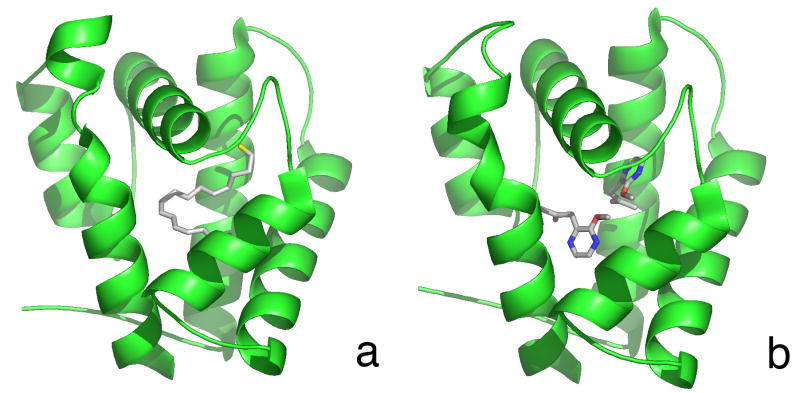
Both the IHD-BmorPBP and BPO-BmorPBP structures (Figs. 3a and b) show an overall protein conformation similar to that of the bombykol-bound structure. The C-terminal region of the protein exists as a disordered loop, and the N-terminal region is an ordered helix. All three disulfide bonds (Leal et al., 1999; Scaloni et al., 1999) are present. In both non-pheromone complexes the protein crystallized with one molecule in the asymmetric unit, rather than as two molecules per asymmetric unit as seen in the bombykol-bound structure, suggesting a subtle change in molecular shape. Previous work using flow injection analysis of the protein followed by mass spectrometry indicated the protein forms a dimer at pH>∼5.5 and a monomer at lower pH (Leal, 2000), though the NMR structure of BmorPBP at physiological pH also shows the protein as a monomer (Lee et al., 2002). The electron density in the pockets of these complexes clearly fits the modeled ligands. Backbone alignments of the proteins were performed using LSQMAN. The root-mean square deviation of the Cα atoms between the iodohexadecane and bombykol complexes was 0.767 Å, and between the bell pepper odorant and bombykol complexes, 0.785 Å, indicating nearly identical backbone conformations among the complexed PBPs (data not shown). The positioning of side chains within the binding pocket is also consistent in all three complexes (Fig. 4). Binding assays indicated that iodohexadecane binds to BmorPBP with significantly lower affinity than the sex pheromone of the silkmoth, bombykol, whereas no binding of the bell pepper odorant was detected (data not shown).
Figure 3. Crystal Structures of BmorPBP in Complex with Nonpheromone Ligands.
Secondary structures are depicted in cartoon representation. Ligands are depicted as ball-and-stick figures. Iodohexadecane complex (A) at 1.9 A ° resolution, and bell pepper odorant complex (B) at 2.0 A ° resolution. Pictures were generated in PyMOL (http://www.pymol.org/).
Figure 4. Alignment of Side Chains Forming the Binding Pocket of BmorPBP.
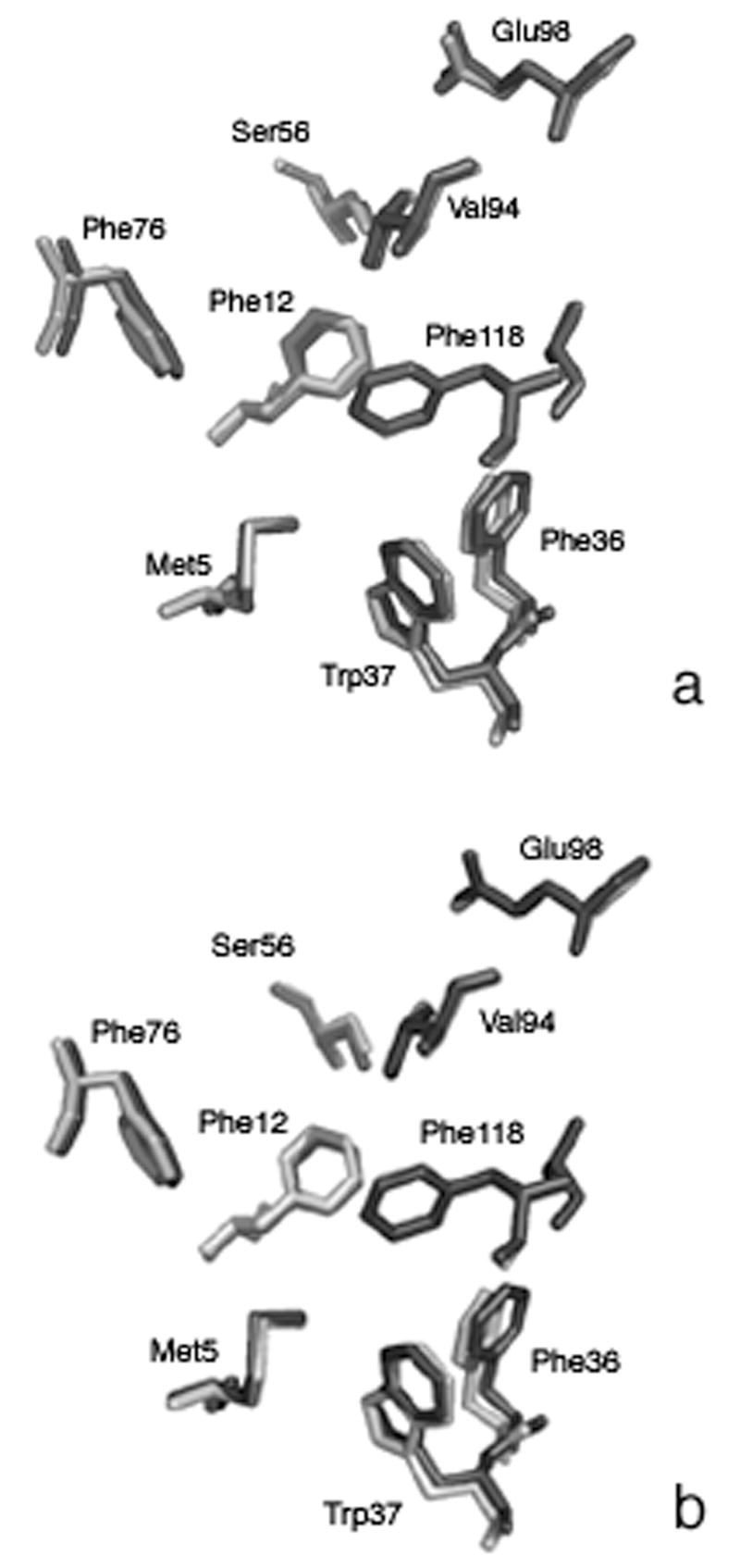
Alignment of bombykol complex binding pocket residues (light gray) and iodohexadecane binding pocket residues (dark gray) in (A). Alignment of bombykol-complex binding pocket residues (light gray) and bell pepper odorant binding pocket residues (dark gray) in (B).
Iodohexadecane complex
The electron density in the binding pocket of the IHD-BmorPBP structure is similar to that of bombykol in the PBP. The electron density of the ligand is continuous (Fig. 5a), and a strong intensity signal presumably representing the electron-rich iodine atom of the ligand was easily identified. The occupancy of all iodohexadecane atoms as defined by a B-factor was less than 50 except for that of the iodine atom, which had a B-factor of 65. The B factors most likely reflect a fairly high occupancy of the ligand within the binding pocket. Although the geometry of iodohexadecane is not restrained by double bonds, the ligand adopted a configuration similar to that of bombykol in the binding pocket. This structure clearly shows the iodine atom of the ligand in closest proximity to Ser56, mimicking the interaction of the alcohol group of bombykol with the Ser56 in the pheromone-PBP structure. In this model, the iodine is 3.7 Å from the sidechain oxygen of Ser56. Other residues located within 4.0 Å of iodohexadecane are Phe12, Leu 62, Leu68, Phe76, Thr111, Val114, Ala115, and Phe118. All these residues also compose the binding pocket in the bombykol complex.
Figure 5. Electron Density Maps of Binding Pocket.
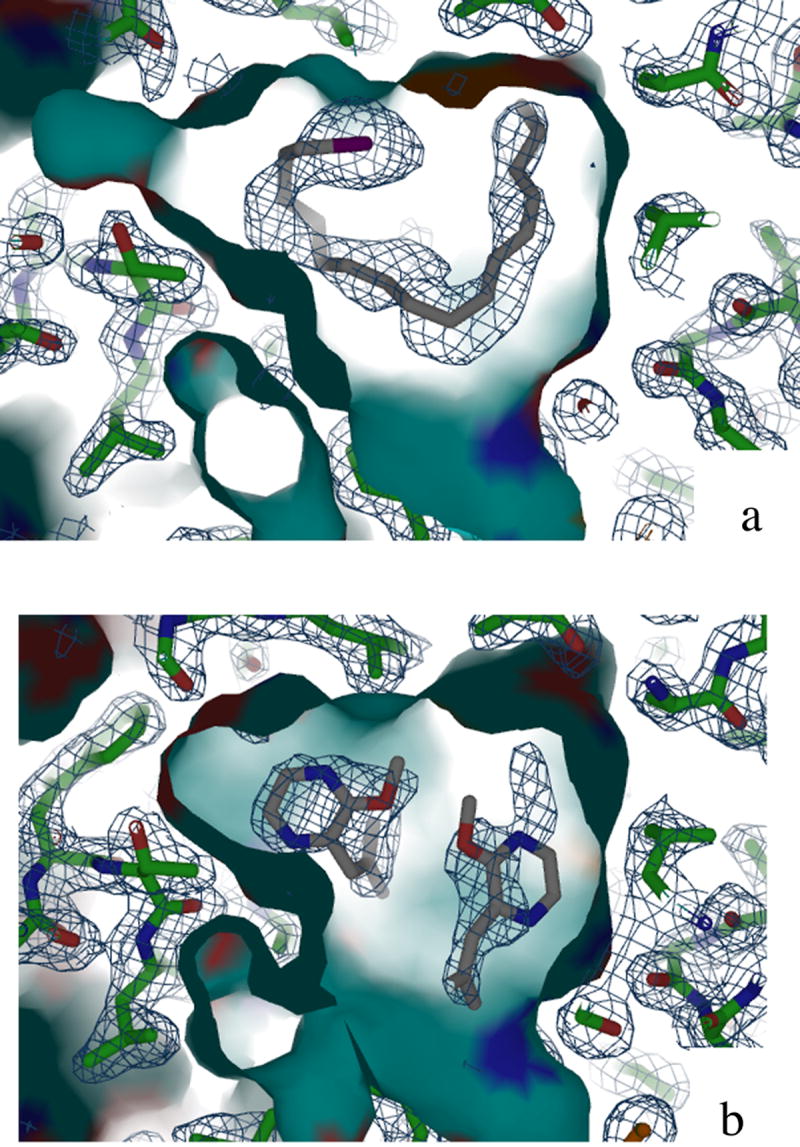
2Fo _ Fc electron density maps of iodohexadecane (A) and bell pepper odorant (B) in BmorPBP binding pocket. A continuous chain of electron density representing all atoms in iodohexadecane is clearly visible in (A). Complete density for one bell pepper odorant molecule and partial density for the second molecule are visible in (B).
Bell pepper odorant complex
Initially, one molecule of bell pepper odorant was modeled into the structure of BPO-BmorPBP. The methoxy- oxygen of bell pepper odorant lays 3.8 Å from the sidechain oxygen of Ser56. The ligand is within at least 4.0 Å of residues Ile52, Ser56, Leu62, Leu68, Val94, Thr111, Val114, Ala115, and Phe118. An additional and comparable mass of electron density was located in the binding pocket, and a second molecule of bell pepper odorant was modeled in (Fig. 5b). Addition of this second model further reduced the Rfree value. These data indicate BmorPBP can accommodate more than one molecule of bell pepper odorant in the binding pocket. The shape of the ring in the second bell pepper odorant molecule is reflected in the electron density, though the density is not complete for this additional molecule. As modeled, this second molecule is within at least 4.0 Å of residues: Leu8, Ser9, Phe12, and Phe36. The closest atom to the methoxy- oxygen of this second bell pepper odorant molecule is Phe12, which is 3.6 Å away. The two bell pepper odorant molecules are separated by a distance of 2.9 Å. All residues in proximity of these two ligand molecules are also involved bombykol-PBP interactions. The occupancy of all bell pepper odorant atoms as defined by a B factor was between 45 and 60, comparable to B factors calculated for bombykol atoms in the pheromone complex. Two water molecules are also modeled into the binding pocket.
Discussion
Based on the structures of these two complexes, B. mori PBP can accommodate diverse hydrophobic molecules within its binding pocket. Polar groups can interact with Ser56, as seen through interactions with the methoxy- group of bell pepper odorant or the iodine of iodohexadecane. Two molecules of bell pepper odorant can be fit into the electron density in the binding pocket of the protein. The structures of these two new complexes show ligands with very different geometries can fit into the cavity of the PBP, from straight chain carbon compounds to aromatic molecules. However, the hook structure adopted by iodohexadecane (similar to the shape of bombykol in the binding pocket) might indicate that appropriate positioning of trans and cis double bonds is influential in binding long chain ligands with restrained geometries. Indeed, the geometry of iodohexadecane in the binding pocket suggests that bombykol, which is locked into this seemingly preferred geometry, is bound over other ligands. Data from the binding assay support this hypothesis. It would be interesting if such “floppy” molecules such as iodohexadecane as used in these crystallography experiments could be used to probe favored ligand geometries in other binding proteins. The discovery that the binding pocket of BmorPBP can accommodate non-pheromone ligands is less surprising given the discovery that the C-terminal tail of the protein occupies the cavity at low pH (Horst et al., 2001) or even at neutral pH when no other ligands are competing for the binding pocket (Oldham et al., 2001). The geometry adopted by iodohexadecane suggests the side chains within the binding pocket of the protein are adapted for the hook-shaped geometry of bombykol, though both iodohexadecane and bell pepper odorant were accommodated by the protein when provided in excess in these crystallographic studies.
Other small binding proteins present in insect antennae, from pheromone-binding proteins to chemosensory proteins, have demonstrated ligand binding flexibility. In the cockroach PBP structure (Lartigue et al., 2003) a fluorescence reporter, amino-naphthalen sulfonate, was bound to the protein, though this molecule was displaced by two out of four components of the species pheromone blend. Even the apo- structure of the same protein contained a glycerol molecule from protein preparation in its binding cavity. One of three PBPs from Antheraea polyphemus was shown to bind with high affinity to all three constituents of the species' pheromonal blend at high pH, but in a competitive assay showed preference for only one component of the blend (Leal et al., 2005a). The structure of a chemosensory protein (CSP) from Mamestra brassicae (Campanacci et al., 2003) showed three molecules of 12-bromo-dodecanol in the binding pocket of the protein, reminiscent of our observation of two bell pepper odorant molecules in the BPO-BmorPBP complex. It is important to note our crystallography experiments are not affinity experiments, and that a 10-fold excess of ligand was incubated with the protein. While iodohexadecane and bell pepper odorant are present in the binding pocket of BmorPBP in these crystal structures, the binding assay did not detect significant binding of these non-pheromone molecules.
The apparent binding flexibility of BmorPBP reflects its affiliation with other antennal odorant-binding proteins. Insect PBPs and OBPs share highly conserved regions, including six conserved cysteine residues that form three disulfide bridges. Both families of proteins have been shown to bind small molecules and shuttle them through the antennal lymph to a receptor. Lepidopteran OBPs show affinities for specific chemical groups or structures, and have been proposed to act as filters to influence which molecules make it to the receptor(s) (Vogt et al., 1991). Recently, it has been demonstrated with transgenic fruit flies expressing either the bombykol receptor only or flies carrying both bombyol receptor and BmorPBP that PBPs are essential for the sensitivity of the insect olfactory system (Syed et al., 2006). In the same way, lepidopteran PBPs may act as filters to speed pheromone processing.
Selectivity and promiscuity of ligand binding to a given lepidopteran PBP could play a role in behavioral antagonism. (Z,Z)-11,13-hexadecadienal is a sexual attractant for both the navel orangeworm, Amyelois transitella Walker, and the meal moth, Pyralis farinalis L. An additional component of the A. transitella pheromone blend, (Z,Z)-11,13-hexadecadien-1-yl acetate, serves as behavioral antagonist for Pyralis farnalis L. (Leal et al., 2005c). It is possible that the PBPs from each species interact differently with the acetate, with P. farnalis PBP binding more strongly and releasing the acetate more slowly, effectually sequestering PBP. Alternatively, a hypothetical multifunctionality of moth PBPs could mimic a routine occurrence in the proteins that develop drug resistance or the ability to degrade chemicals, where proteins have adapted to function differently in the presence of a new and sometimes strikingly different chemical. At least modest PBP ligand promiscuity would be useful to facilitate speciation in moths (Roelofs et al., 2002).
These studies suggest BmorPBP is capable of binding non-pheromone ligands in the moth antenna, though with affinity lower than that detected by the binding assay. Under physiological conditions, pheromone binding proteins greatly outnumber any potential ligand, and the apparent promiscuity of BmorPBP indicated by crystallographic studies may be a reflection a protein that has evolved to pick up anything resembling a pheromone signal. That BmPBP can bind very different ligands with only minor side chain adjustments argues that it has evolved for plasticity. Non-pheromone ligand might be dropped by the protein en route to the receptor, as only bombykol was shown to bind significantly in binding assays. In this way, the protein would act as a filter to decrease the number of small molecules that are presented to the receptor while maintaining sensitivity, increasing the overall speed of pheromone processing. Ligands with low affinity may be dropped from the complex and inactivated by aggressive odorant-degrading enzymes (Ishida and Leal, 2005; Vogt et al., 1985) while en route to the receptors, whereas pheromone molecules may remain protected by PBPs until the end of the journey. The gain in sensitivity may offset the loss in selectivity, particularly because of the additional layer of selectivity provided by the receptors.
Experimental Procedures
Protein Preparation
Recombinant BmorPBP protein was expressed in E. coli BL21 (DE3) cells and purified as previously described (Wojtasek and Leal, 1999). A slight modification of the previous purification procedure was used to remove any adventitious hydrophobic ligands the protein might have picked up during expression (Oldham et al., 2001). Recombinant PBP was dialyzed against 0.2 M sodium phosphate buffer pH 4.5 and incubated with a Lipidex resin at 37 °C to remove any hydrophobic molecule acquired during recombinant expression. No ligand is found in the crystal structure of the unliganded protein (Lautenschlager et al., 2005) when using this protocol, ensuring the only molecules present for binding are those added to the protein. Ligand-free protein was eluted with 0.2 M sodium phosphate pH 4.5.
Ligand addition
Ligands were dissolved in methanol to make them more soluble for protein complex formation. BmorPBP concentration was determined by Bradford Assay, and ligand was added to the protein in a ten-fold molar excess. The protein-ligand mixture was agitated briefly by vortex and incubated at 4 °C overnight, then the protein complex was dialyzed against 10 mM Tris buffer pH 8.0 in Fisherbrand dialysis tubing (MWCO: 6-8 kDa). Protein complexes were concentrated to 20 mg/ml in an Amicon Ultra-15 Centrifugal Filter Unit (NMWL: 10 kDa), and unbound ligand removed by centrifugation on at 13,000x g for 5 minutes at 4 °C. This same procedure was used to purify, crystallize, and re-solve the BmorPBP-bombykol complex as a control, following the above procedure prior to crystallization to ensure the validity of the method.
Crystallization and Structure Determination
Crystals were obtained by vapor diffusion hanging drop method at 22 °C. Drops containing 2 μl protein complex in 10 mM Tris pH 8.0 and 2 μL of well solution containing 40% polyethylene glycol (MW 4,000), 100 mM Tris pH 8.0, and 50 mM MgCl2 were set up in hanging drop trays. Complete data sets at 1.9 Å resolution for IHD-BmorPBP and 2.0 Å resolution for the BPO-BmorPBP were collected on F1 at CHESS (Cornell University, Ithaca, NY). The data were integrated using DENZO of the HKL suite (Otwinowski, 1997) followed by scaling with SCALA of the CCP4 suite (Collaborative Computation Project, 1994). Data set statistics are shown in Table 1. Further data reduction was performed with CCP4i.
The structures were solved by molecular replacement using the CCP4 version of MOLREP (Vagin and Teplyakov, 2000). The model used was the B. mori PBP bombykol complex, chain A (Protein Data Bank entry 1DQE). Model building was done with O (Jones et al., 1991). The structure was completed and refined by iterative cycles of model building and simulated annealing using O and CNS v.1.1 (Brunger et al., 1998). The eight C-terminal residues were omitted in the final refinement of each complex due to structural disorder.
Binding Assays
Binding was measured by incubating BmorPBP with test ligands, separating unbound and bound ligand, extracting bound ligrand from the protein, and quantifying the amount of bound ligand by gas chromatography, according to a previously reported protocol (Leal et al., 2005b).
Acknowledgments
This work is based upon research conducted at the Cornell High Energy Synchrotron Source (CHESS) which is supported by the National Science Foundation and the National Institutes of Health/National Institute of General Medical Sciences under award DMR 9713424. Work supported by NIH CA59021 (JC), NIH 1U01AI058267-01, USDA 2003-35302-13648, NSF 0234769 (WL).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
ACCESSION NUMBERS The iodohexadecane-BmorPBP complex has been deposited with RCSB ID 042046 and PDB ID number 2P71. The bell pepper odorant-BmorPBP complex has been deposited with RCSB ID 042045 and PDB ID number 2P70.
References
- Ando T, Inomata SI, Yamamoto M. Lepidopteran sex pheromones. Top Curr Chem. 2004;239:51–96. doi: 10.1007/b95449. [DOI] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta crystallographica. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Butenandt A. Bombykol, the sex attractive substance of the silkworm moth Bombyx mori. Endocrinology. 1963;27:9. [Google Scholar]
- Campanacci V, Lartigue A, Hallberg BM, Jones TA, Giudici-Orticoni MT, Tegoni M, Cambillau C. Moth chemosensory protein exhibits drastic conformational changes and cooperativity on ligand binding. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:5069–5074. doi: 10.1073/pnas.0836654100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collaborative Computation Project, N. The CCP4 suite: programs for protein crystallography. Acta crystallographica. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- Horst R, Damberger F, Luginbuhl P, Guntert P, Peng G, Nikonova L, Leal WS, Wuthrich K. NMR structure reveals intramolecular regulation mechanism for pheromone binding and release. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:14374–14379. doi: 10.1073/pnas.251532998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida Y, Leal WS. Rapid inactivation of a moth pheromone. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:14075–14079. doi: 10.1073/pnas.0505340102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991;47(Pt 2):110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- Lartigue A, Gruez A, Spinelli S, Riviere S, Brossut R, Tegoni M, Cambillau C. The crystal structure of a cockroach pheromone-binding protein suggests a new ligand binding and release mechanism. The Journal of biological chemistry. 2003;278:30213–30218. doi: 10.1074/jbc.M304688200. [DOI] [PubMed] [Google Scholar]
- Lautenschlager C, Leal WS, Clardy J. Coil-to-helix transition and ligand release of Bombyx mori pheromone-binding protein. Biochemical and biophysical research communications. 2005;335:1044–1050. doi: 10.1016/j.bbrc.2005.07.176. [DOI] [PubMed] [Google Scholar]
- Leal WS. Duality monomer-dimer of the pheromone-binding protein from Bombyx mori. Biochemical and biophysical research communications. 2000;268:521–529. doi: 10.1006/bbrc.2000.2158. [DOI] [PubMed] [Google Scholar]
- Leal WS. Proteins that make sense. In: Blomquist GJ, Vogt RG, editors. Insect Phermone Biochemistry and Molecular Biology. Elsevier Academic Press; 2003. pp. 447–467. [Google Scholar]
- Leal WS. Pheromone reception. Top Curr Chem. 2005;240:1–36. [Google Scholar]
- Leal WS, Chen AM, Erickson ML. Selective and pH-dependent binding of a moth pheromone to a pheromone-binding protein. Journal of chemical ecology. 2005a;31:2493–2499. doi: 10.1007/s10886-005-7458-4. [DOI] [PubMed] [Google Scholar]
- Leal WS, Chen AM, Ishida Y, Chiang VP, Erickson ML, Morgan TI, Tsuruda JM. Kinetics and molecular properties of pheromone binding and release. Proceedings of the National Academy of Sciences of the United States of America. 2005b;102:5386–5391. doi: 10.1073/pnas.0501447102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leal WS, Nikonova L, Peng G. Disulfide structure of the pheromone binding protein from the silkworm moth, Bombyx mori. FEBS letters. 1999;464:85–90. doi: 10.1016/s0014-5793(99)01683-x. [DOI] [PubMed] [Google Scholar]
- Leal WS, Parra-Pedrazzoli AL, Kaissling KE, Morgan TI, Zalom FG, Pesak DJ, Dundulis EA, Burks CS, Higbee BS. Unusual pheromone chemistry in the navel orangeworm: novel sex attractants and a behavioral antagonist. Die Naturwissenschaften. 2005c;92:139–146. doi: 10.1007/s00114-004-0598-5. [DOI] [PubMed] [Google Scholar]
- Lee D, Damberger FF, Peng G, Horst R, Guntert P, Nikonova L, Leal WS, Wuthrich K. NMR structure of the unliganded Bombyx mori pheromone-binding protein at physiological pH. FEBS letters. 2002;531:314–318. doi: 10.1016/s0014-5793(02)03548-2. [DOI] [PubMed] [Google Scholar]
- Maida R, Ziegelberger G, Kaissling KE. Ligand binding to six recombinant pheromone-binding proteins of Antheraea polyphemus and Antheraea pernyi. Journal of comparative physiology. 2003;173:565–573. doi: 10.1007/s00360-003-0366-4. [DOI] [PubMed] [Google Scholar]
- Mohanty S, Zubkov S, Gronenborn AM. The solution NMR structure of Antheraea polyphemus PBP provides new insight into pheromone recognition by pheromone-binding proteins. Journal of molecular biology. 2004;337:443–451. doi: 10.1016/j.jmb.2004.01.009. [DOI] [PubMed] [Google Scholar]
- Oldham NJ, Krieger J, Breer H, Svatos A. Detection and removal of an artefact fatty acid from the binding site of recombinant Bombyx mori pheromone-binding protein. Chemical senses. 2001;26:529–531. doi: 10.1093/chemse/26.5.529. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Pelosi P. Odorant-binding proteins. Critical reviews in biochemistry and molecular biology. 1994;29:199–228. doi: 10.3109/10409239409086801. [DOI] [PubMed] [Google Scholar]
- Prestwich GD, Du G. Pheromone-binding proteins, pheromone recognition, and signal transduction in moth olfaction. In: Carde RT, Minks AK, editors. Insect Pheromone Research. New York: Chapman and Hall; 1997. [Google Scholar]
- Roelofs WL, Liu W, Hao G, Jiao H, Rooney AP, Linn CE., Jr Evolution of moth sex pheromones via ancestral genes. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:13621–13626. doi: 10.1073/pnas.152445399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandler BH, Nikonova L, Leal WS, Clardy J. Sexual attraction in the silkworm moth: structure of the pheromone-binding-protein-bombykol complex. Chemistry & biology. 2000;7:143–151. doi: 10.1016/s1074-5521(00)00078-8. [DOI] [PubMed] [Google Scholar]
- Scaloni A, Monti M, Angeli S, Pelosi P. Structural analysis and disulfide-bridge pairing of two odorant-binding proteins from Bombyx mori. Biochemical and biophysical research communications. 1999;266:386–391. doi: 10.1006/bbrc.1999.1791. [DOI] [PubMed] [Google Scholar]
- Syed Z, Ishida Y, Taylor K, Kimbrell DA, Leal WS. Pheromone reception in fruit flies expressing a moth's odorant receptor. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:16538–16543. doi: 10.1073/pnas.0607874103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagin A, Teplyakov A. An approach to multi-copy search in molecular replacement. Acta crystallographica. 2000;56:1622–1624. doi: 10.1107/s0907444900013780. [DOI] [PubMed] [Google Scholar]
- Vogt RG. Pheromone Biochemistry. In: Prestwich GD, Blomquist GJ, editors. Pheromone Biochemistry. Orlando: Academic Press, Inc; 1987. pp. 385–431. [Google Scholar]
- Vogt RG, Prestwich GD, Lerner MR. Odorant-binding-protein subfamilies associate with distinct classes of olfactory receptor neurons in insects. Journal of neurobiology. 1991;22:74–84. doi: 10.1002/neu.480220108. [DOI] [PubMed] [Google Scholar]
- Vogt RG, Riddiford LM, Prestwich GD. Kinetic properties of a sex pheromone-degrading enzyme: the sensillar esterase of Antheraea polyphemus. Proceedings of the National Academy of Sciences of the United States of America. 1985;82:8827–8831. doi: 10.1073/pnas.82.24.8827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojtasek H, Leal WS. Conformational change in the pheromone-binding protein from Bombyx mori induced by pH and by interaction with membranes. The Journal of biological chemistry. 1999;274:30950–30956. doi: 10.1074/jbc.274.43.30950. [DOI] [PubMed] [Google Scholar]