Abstract
Many human neurodegenerative diseases are associated with amyloid fibril formation. The human 99-residue β2-microglobulin (β2m) is one of the most intensively studied amyloid-forming proteins. Recent studies show that the C-terminal fragments 72–99, 83–89, and 91–96 form by themselves amyloid fibrils in vitro and play a significant role in fibrillization of the full-length β2m protein under acidic pH conditions. In this work, we have studied the equilibrium structures of the 17-residue fragment 83–99 in solution, and investigated its dimerization process by multiple molecular dynamics simulations. We find that an intertwined dimer, with the positions of the β-strands consistent with the results for the monomer, is a possible structure for two β2m(83–89) peptides. Based on our molecular-dynamics-generated dimeric structure, a protofibril model is proposed for the full-length β2m protein.
INTRODUCTION
More than 20 human diseases, including Alzheimer's disease, prion diseases, and dialysis-related amyloidosis, are associated with the pathological self-assembly of soluble proteins into cytotoxic oligomers and amyloid fibrils (1–3). Although the proteins share very little sequence identity and structure similarity in solution, x-ray diffraction data show that all fibrils contain a common cross β-sheet structure with β-strands perpendicular and hydrogen bonds parallel to the fibril axis (4), suggesting a common aggregation mechanism for all amyloidogenic proteins. Understanding the detailed aggregation paths is an important step toward rational design of improved therapeutics preventing neurodegenerative diseases. However, due to the transient nature of oligomers, the structures of the intermediate species are poorly understood at an atomic level of detail.
In dialysis-related amyloidosis, the amyloidogenic component is the 99-residue human β2-microglobulin (β2m) protein (5), the light chain of the class I major histocompatibility complex (MHC-I). After its release from the cells expressing MHC-I, β2m is carried to the kidney where it is degraded (6). When renal failure occurs, usually causing high concentration of β2m, the soluble protein aggregates into insoluble amyloid fibrils (7). Deposition of β2m fibrils is a serious complication in patients who have been subjected to long-term hemodialysis (8). The β2m fibrils are often deposited on synovial membranes around large joints, and therefore this type of amyloidosis characteristically induces severe bone/joint complications. It is well established that β2m forms amyloid fibrils under acidic pH conditions (9–11), and that the morphology of the fibrils is pH-dependent (12).
Several molecular dynamics (MD) simulations on full-length β2m and its amyloid-forming fragments have been conducted. Two distinct β2m intermediates, suggested to be on the paths to amyloid fibril, were observed by pH-mediated partial unfolding MD simulations at 310 K and temperature-mediated unfolding MD simulations at 498 K (13). The role of the single Cys25-Cys80 disulfide bond on β2m aggregation was studied by discrete MD (14). The structural properties of the fragments β2m(21–31) and β2m(83–89) were also determined in explicit water. The β2m(21–31) monomer was found to adopt a β-hairpin using a 4.3 ns replica exchange molecular dynamics (REMD) simulation (15). The β2m(83–89) dimer was studied by multiple 50-ns MD simulations at 310 K with main-chain conformational restraints, and both parallel and antiparallel β-sheets were observed (16).
Although β2m has been known as a human amyloid protein for more than 20 years, the molecular mechanisms underlying the self-assembly of β2m are not fully understood. Several critical regions for amyloid fibril formation have been identified (17–19) and one of these regions spans the C-terminal amino acids 72–99 (20). Besides the region 72–99, it has also been found that the fragments 83–89 and 91–96 can also form by themselves amyloid fibrils in vitro (21,22). The importance of the C-terminal region in the context of β2m fibril formation is also supported by the observation that binding of a monoclonal antibody to residues 92–99 inhibits amyloid formation, while binding to residues 20–41 or 63–75 has no effect on β2m fibrillogenesis (23). Very recently, based on the experimental observation that the fragment 83–89 from human β2m forms amyloid fibrils, Eisenberg et al. proposed a fibrillar model for the full-length β2m protein, in which the amino acids 83–99 form the spine of the protofibrils (21,24).
Considering the role of the C-terminal amino acids in β2m fibril formation as revealed by experiments (20,21,23) and the role of residues 83–99 in the Eisenberg's β2m fibril (21), we choose the C-terminal 17-residue β2m(83–99) peptide for study. In this work, we perform two series of molecular dynamics (MD) simulations in explicit water at acid pH. First, we investigate in detail the equilibrium structures of the β2m(83–99) monomer by using three independent REMD simulations (totaling 4.5 μs). Second, we run six independent MD trajectories (totaling 0.815 μs) to explore the self-assembly and the resulting aggregates of a dimer at 310 K. We find that, although the β2m(83–99) monomer is essentially random coil in water, the N-terminal residues 86–89 and the C-terminal residues 95–98 have a high intrinsic propensity to adopt aggregation-prone β-strands. We also show that two β2m(83–99) peptides aggregate into a β-sheet structure with two interlaced hairpins. Based on this structure, we construct a protofibril model for β2m.
MATERIALS AND METHODS
The sequence of the β2m(83–99) peptide is NHVTLSQPKIVKWDRDM. A summary of the setup details of all simulations is given in Table 1. For each system, we describe the method used, the label of the runs, the protonation state of the termini, the simulation time, the temperature at which simulations were performed, and the starting conformation. As seen, we use two protonation states of the peptide.
TABLE 1.
Setup details of all simulations
| System | Method | Label | Protonation state* | Time | Temperature | Initial States |
|---|---|---|---|---|---|---|
| Monomer | REMD | REMD1 | NH2, COO− | 25 | 300 ∼ 520 | Fig. S1 a |
| Monomer | REMD | REMD2 | NH2, COO− | 25 | 300 ∼ 520 | Fig. S1 a |
| Monomer | REMD | REMD3 | NH3+, COOH | 25 | 300 ∼ 520 | Fig. S1 a |
| Dimer | MD | Runs A–C | NH2, COO− | 200 × 3 | 310 | Fig. 4 b |
| Dimer | MD | Run D | NH2, COO− | 40 | 310 | Fig. S2 a |
| Dimer | MD | Run E | NH3+, COOH | 40 | 310 | Fig. S2 b |
| Dimer | MD | Run F | NH3+, COOH | 60 | 310 | Fig. S2 c |
| C-dimer | MD | R1 ∼ 15 | NH2, COO− | 5 × 15 | 310 | Fig. 6 |
| Protofibril | MD | N/A | NH2, COO− | 20 | 310 | Fig. 8 a |
| Protofibril | MD | N/A | NH2, COO− | 10 | 330 | Fig. 8 a |
For each system, we describe the method used, the label of the runs, the protonation state of the termini, the simulation time (in nanoseconds), the temperature (K), and the starting states.
For all species, the protonation state of the side chains of Lys+, Arg+, His+, and Asp0 is kept constant.
To mimic experimental pH ranging from 2.6 to 3.9, the side chains of Lys, Arg, His, and Asp are protonated (Lys+, Arg+, His+, Asp0), and the C-terminus is deprotonated (COO−) (25). The N-terminus is also taken as deprotonated (NH2), while it should be protonated, to mimic its protonation state within the full protein. The net charge of the peptide with NH2 and COO− termini is +3. To mimic pH values smaller than 2.6, we use the protonated NH3+ and COOH termini, leading to a +5 net charge on the peptide. Water is modeled by the explicit simple-point charge model (26).
β2m(83–99) monomer
The initial conformation is taken from the residues 83–99 of human β2-microglobulin (PDB code: 1JNJ): an extended conformation with residues 91–94 in β-strand and the other residues in random coil. The minimum distance between the monomer and the water box wall is 1.2 nm. The size of the monomer system (peptide and water) is 13,833 atoms. A 1000-step steepest descent minimization and 100-ps MD equilibration are performed before the production run. After equilibration, the conformation is used for the REMD run (REMD1) and the dimer MD runs. This conformation shown in Supplementary Material Fig. S1 a is essentially extended with bends for residues 83–87 and coils for the other residues.
Two distinct conformations generated by a MD run at 450 K and shown in Supplementary Material Fig. S1 a are used as the starting points of the REMD2 and REMD3 runs. The REMD1 and REMD2 runs sharing the NH2 and COO− termini are used to check the convergence of the sampling. In contrast, the REMD3 run with the NH3+ and COOH termini is used to examine the effect of the protonation state of peptide termini on the equilibrium structures. All simulations are performed using periodic boundary conditions in a truncated octahedron box.
β2m(83–99) dimer
For the 200-ns MD runs (Runs A–C), the peptides were put 2.25 nm apart, as monitored by the distance of their centers of mass, and in a random orientation (see the structure in Fig. 4 b at t = 0 ns). The minimum distance between the dimer and the water box wall is 1.0 nm. For the short trial MD runs (5 ns each), the initial structure is described in Discussion and Conclusions. There are 24,723 atoms in total (peptides and water).
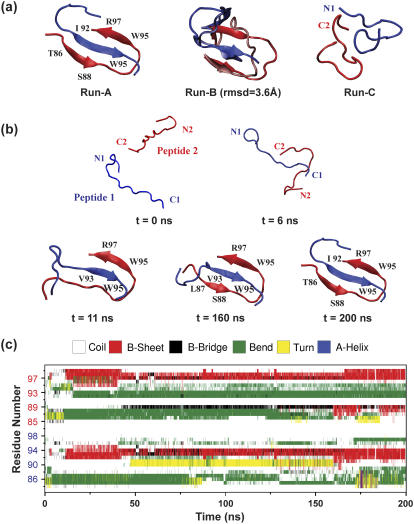
FIGURE 4.
Assembly of β2m(83–99) dimer. (a) Final 200-ns structures generated by Run A (left) and Run C (right), along with the superposition of the structures at 58 and 200 ns (middle) in Run B. (b) Five representative snapshots at 0, 6, 11, 160, and 200 ns of Run A. The two peptides are in blue and red. (c) Time evolution of secondary structure in Run A using the DSSP program (30).
For the two 40-ns MD runs (Runs D and E), the initial structure consists of a preformed anti-parallel β-sheet spanning residues V93-R97 and coils for the other residues (Supplementary Material Fig. S2, a and b. The protonation state of peptide termini is varied. Runs D and E use a net charges of +3 and +5 for each chain, respectively.
For the 60-ns MD run (Run F), the initial structure consists of a preformed anti-parallel β-sheet (the same as the preformed anti-parallel β-sheet in the initial structure of Run E) and a fully extended state for the other residues (Supplementary Material Fig. S2 c). Here, each peptide has a net charge of +5.
All simulations are performed using periodic boundary conditions in a truncated octahedron box.
β2m(83–99) protofibril
Two MD simulations (20 ns at 310 K and 10 ns at 330 K) are performed using periodic boundary conditions in a rectangular water box. Here each peptide has a +3 net charge. The protofibril model was constructed by two β-sheet layers with antiparallel arrangement. The initial distance between the two sheets is set to 1.4 nm. The entire system has a total of 47,400 atoms. The box dimensions are 7 nm × 7 nm × 10 nm.
MD and REMD simulations
The MD simulations are performed using the GROMACS software package (27) and GROMOS96 force field (28). The LINCS and SETTLE algorithms are used to constrain all bond lengths of the peptides and water molecules, respectively, allowing an integration time step of 2 fs. A twin-range cutoff 1.0/1.4 nm is used for the nonbonded interactions, and a reaction-field correction with dielectric permittivity ε = 80 is employed to calculate long-range electrostatics interactions. The temperature and the pressure are maintained by coupling temperature and pressure baths using Berendsen's method (29). Although the Berendsen temperature coupling does not strictly reproduce the canonical distribution, it is a widely used thermostat due to its stability and efficiency in MD simulations. The solute and solvent are separately coupled to external temperature and pressure baths. The temperature-coupling constant is 0.1 ps. The pressure is kept at 1 bar using weak pressure coupling with τp = 1.0 ps (29).
The REMD simulations are carried out in the NVT ensemble. Each REMD simulation uses 60 replicas of 25 ns in parallel. The temperature ranges from 300 K to 520 K with exponential distribution. The swap time between neighboring replicas is 1 ps. The acceptance ratio is between 20% and 38%.
All trajectories are analyzed with the facilities implemented in GROMACS software package (27). In the analysis of REMD runs, the first two-nanosecond trajectories were excluded. The secondary structure content was identified using the DSSP program (30).
RESULTS
Structure and potential of mean force of β2m(83–99) monomer
To characterize the structure of the monomer in a pH range 2.6–3.9, we perform two REMD simulations starting from distinct conformations. REMD convergence is checked by comparing the β-sheet probability of each residue generated by REMD1 and REMD2 at 310 K. It can be seen from Supplementary Material Fig. S1 that the β-sheet probability profiles superpose very well, but display deviations of 6% at positions 96 and 98, indicating a reasonable convergence of our simulations. Analysis of the structures shows, however, that the same main conformational space are sampled by the REMD1 and REMD2 simulations with differences in Boltzmann probabilities of the dominant states reaching at most 4% at 310 K.
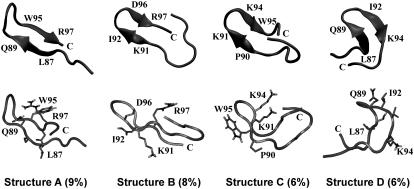
The REMD1 and REMD2 data are combined for analysis. All the conformations at 310 K can be classified into five families. The monomer is essentially random coil (71% population, structures not shown). However, it also adopts multiple β-hairpin-like conformations (29% population), differing in the positions of the β-sheets and turns (including 3-, 4-, and 5-type turns as identified by DSSP program). A representative conformation of each β-hairpin (with its population) is shown in Fig. 1.
FIGURE 1.
REMD-generated hairpin-like structures of β2m(83–99) monomer at 310 K. Their populations are given in parentheses. The position of the C-terminal end is indicated. Each structure is shown with the location of the strands (top panel) and the location of selected side-chains (bottom panel).
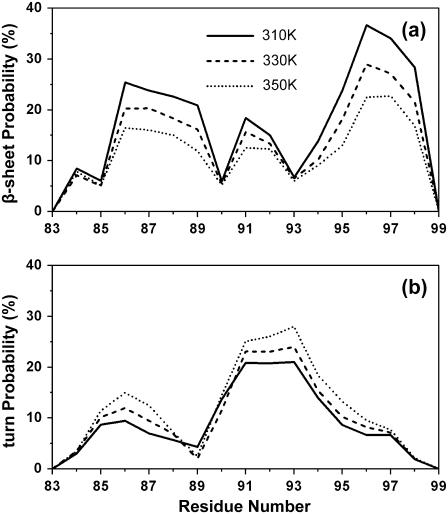
The secondary structure (β-sheet and turn) probability of each residue is shown in Fig. 2 at 310, 330, and 350 K. It can be seen from Fig. 2 a that, at 310 K, there are two dominant regions with high β-sheet content: region A (residues 86–89, 21–25% of the time) and region B (residues 95–98, 24–34% of the time). The region B, located in the C-terminal, has thus a higher propensity for β-sheet than the region A located in the N-terminal. Fig. 2 b shows that residues 91–93 spent ∼21% of their time in turn conformation at 310 K. The β-sheet content decreases gradually with increasing temperature, while the turn content is stable at high temperature. This temperature-dependent behavior of turn content has also been observed for the amyloid fragments Aβ(25–35) and Aβ(12–28) by REMD (31,32). Moreover, it has been noted that the amount of turn content present in Aβ(25–35) is positively correlated with the ability of the peptide to aggregate (33). We know that the β-sheet probabilities using GROMOS96 might be quantitatively different using another force field. Using two peptides with α-helical and β-hairpin character, Yoda et al. found that GROMOS96 has a bias toward β conformations, while OPLS has not (34). Recent studies on Aβ(25–35) (31) and Aβ(10–35) peptides (35) show, however, very similar results using GROMOS96 and OPLS force fields. The effects of the force field on the structure of β2m(83–99) peptide remains therefore to be determined, but such a study is beyond the scope of this work.
FIGURE 2.
Secondary structure probability of each residue within the monomer determined by DSSP (30) at 310, 330, and 350 K: (a) β-sheet probability; (b) β-turn probability.
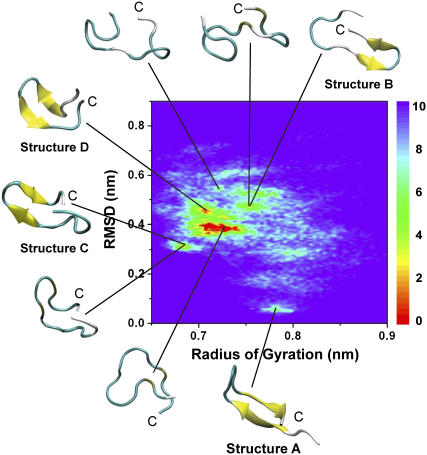
The potential of mean force at 310 K is plotted in Fig. 3 as a function of the radius of gyration (rg) and the main-chain root mean-square deviation (RMSD) from the structure A shown in Fig. 1. Representative conformations of low free energies are also shown. Four basins with β-hairpin character (structures A–D in Fig. 1) are located at (rg, RMSD) values of (0.73 nm, 0.38 nm), (0.71 nm, 0.46 nm), (0.78 nm, 0.05 nm), and (0.68 nm, 0.31 nm), respectively. In contrast, various random coils are distributed in the whole free energy map.
FIGURE 3.
Potential of mean force of β2m(83–99) monomer at 310 K. The potential of mean force is plotted as a function of the radius of gyration (rg) and the RMSD from structure A shown in Fig. 1. Representative structures of low free energies (in kJ/mol) are shown.
To determine the effect of the NH3+ and COOH termini on the equilibrium structures at pH 2.6 or less, we performed a 25-ns REMD3 simulation starting from Supplementary Material Fig. S1 a. In Supplementary Material Fig. S1 b, we see that the β-sheet probabilities of each residue from REMD1, REMD2, and REMD3 are very similar, with deviations of 6% at positions 96 and 98. Structural analysis of the dominant states from the three REMD runs shows, however, that the same main conformational space is sampled, although the protonation states of peptide termini are different. This indicates a small effect of protonation state of the N- and C-termini on the equilibrium structures of the β2m(83–99) monomer.
Assembly of β2m(83–99) dimer
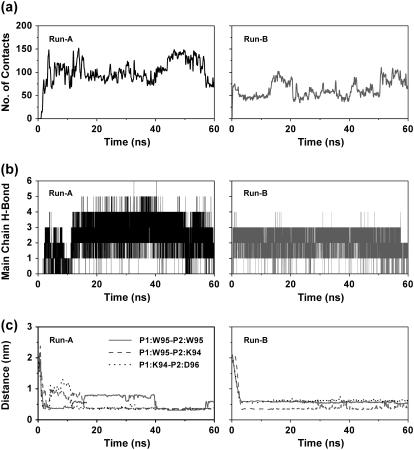
To explore the assembly of two β2m(83–99) peptides, three independent MD simulations (200 ns each) are performed in explicit water at 310 K and pH ranges from 2.6 to 3.9, starting from a conformation described in Materials and Methods and shown in Fig. 4 b. Two runs (Run A and Run B) locate two interlaced β-hairpins, while Run C displays a disordered aggregate within a 200-ns timescale (Fig. 4 a). For the Run B, we also show the superposition of the structure generated at 200 ns on the structure generated at 58 ns. These structures deviate by 3.6 Å RMSD from each other.
The dimerization process observed in Run A is described in Fig. 4, b and c). Starting from an initial state without any intermolecular contacts between the C-terminal regions and between the N-terminal regions, the C-terminal ends come in proximity very rapidly (at t < 6 ns, Fig. 4 b). At this point, the N-terminal residues V85-L87 adopt bend conformations, while the other residues are random coil (Fig. 4 c). At t = 11 ns, a random coil to β-strand transition occurs within the C-terminal residues and an anti-parallel β-sheet forms between residues V93-W95 of peptide 1 (referred to as P1) and residues W95-R97 of peptide 2 (P2) (see Fig. 4 b). Subsequently, the N-terminal region of P2 folds over to align with the C-terminal region of P1. At t = 48 ns, the residue P2:S88 adopts a β-bridge (Fig. 4 c). Afterwards, its neighboring residues T86 and L87 adopt β-bridge conformations successively. At t = 160 ns, a parallel β-sheet forms between the C-terminal residues V93-K94 of P1 and the N-terminal residues L87-S88 of P2. During the last 40 ns of the simulation, the two previous strands are extended and lead to a longer parallel β-sheet involving P1:I92-K94 and P2:T86-S88. At t = 200 ns, we see that the N-terminal region of P1 has folded back on the C-terminal region of P2 and the dimer is assembled into two interlaced hairpins.
Although the details of the side-chain contacts and hydrogen-bonding networks differ from the two final structures in Runs A and B (Fig. 4 a), both runs share the same assembly process: the C-terminal regions of P1 and P2 come first in proximity and form an anti-parallel β-sheet, afterwards the N-terminal region of P2 folds over to align with the C-terminal region of P1 and self-organizes to a parallel β-sheet, finally an interlaced-hairpin-like β-sheet dimer forms.
To monitor the early formation of the antiparallel β-sheet between the peptides P1 and P2 in Runs A and B, we plot the time evolution of the total number of sidechain-sidechain contacts (Fig. 5 a) and number of main-chain H-bonds (Fig. 5 b) between the interpeptide C-terminal amino acids (residues I92–R97) within the first 60 ns. Here, we consider that two interpeptide heavy atoms are in contact if their distances come within their van der Waals radii + 2.8 Å (36), and a H-bond is formed if the donor-acceptor distance is <3.5 Å and the donor-hydrogen-acceptor angle >150°. As seen in Fig. 5 a, the number of interpeptide contacts between the C-terminal regions increases very rapidly within the first 2 ns, and then fluctuates at ∼125 ± 25 and 75 ± 25 in Runs A and B, respectively. Along with these sidechain-sidechain interactions, we also see that the H-bonds form within 2 and 1 ns in Runs A and B, respectively.
FIGURE 5.
Analysis of the formation of the antiparallel β-sheet between the interpeptide C-terminal amino acids (residues I92-R97) within the first 60 ns of the Runs A and B. Time evolution of (a) the total number of sidechain-sidechain contacts; (b) the total number of H-bonds; and (c) the minimum distance between three pairs of side chains: P1:W95-P2:W95, P1:W95-P2:K94, and P1:K94-P2:D96.
To elucidate which interactions play a key role in the formation process of the interpeptide antiparallel β-sheet, Fig. 5 c shows the time evolution of the minimum distance between three pairs of side-chains: P1:W95-P2:W95, P1:W95-P2:K94, P1:K94-P2:D96, in the first 60 ns of the simulations. The expression P1:Xi-P2:Yj denotes the interaction between residue Xi of P1 and residue Yj of P2. Initially, all distances are >1.5 nm. In both runs, the three pairs of side chains come within 6.0 Å at 4 ns. Subsequently in Run A, the minimum distances P1:W95-P2:K94 and P1:K94-P2:D96 increase momentarily to 1.0 nm between 4 and 16 ns, but they decrease again after 16 ns. This indicates that the formation and stabilization of the β-sheet results from aromatic-aromatic interactions (W95-W95), electrostatic interactions (K94-D96), and the interaction between W95 and the hydrophobic component of K94.
DISCUSSION AND CONCLUSIONS
The structures of β2m(83–99) monomer have been investigated in explicit solvent by using two independent REMD simulations at a pH range from 2.6 to 3.9 and one REMD simulation at a pH of 2.6 or less. We find that at 310 K the peptide is predominantly random coil. This is consistent with the NMR-derived dynamics of the unfolded state of full-length β2m, where residues 85–99 were observed to be highly dynamic and closely represented by a random coil conformation (37). Our prediction of a low population of β-hairpin for the monomer is also of interest, since the β-sheet propensity of the residues contributes to amyloid formation (38,39). Our simulations point to two regions: region A spanning residues 86–89 and region B spanning 95–98, and a higher probability of the region B for β-sheet. This is fully consistent with ab initio prediction using the CSSP method (40).
Our 200-ns dimer simulations with the NH2 and COO− termini show that two runs, driven by interpeptide C-terminal amino acids interactions (referred to as CC), lead to β-structure, while an amorphous structure is reached by early NC interactions (data not shown). To determine whether interpeptide CC or NC interactions are more likely to drive the early steps of aggregation, we repeat 15 short trial MD simulations (5 ns each) at 310 K starting from a C-shape structure, shown in Fig. 6, where all the Cα-Cα distances between the interpeptide termini are identical and set to 1.9 nm. This construction allows us therefore to examine the competition between various scenarios, namely early formation of interpeptide NN, CC, and NC interactions. Here, the N-terminal region spans residues H84–Q89, and residues I92–R97 still represent the C-terminal region.
FIGURE 6.
Two different views of the initial state of β2m(83–99) dimer used for the 5-ns trial MD simulations. Both monomers have a C-shape structure. The Cα-Cα distance between N1 and N2, N1 and C2, N2 and C1, and C1 and C2 termini is identical and equal to 1.9 nm.
Table 2 gives the time for first occurrence of sidechain-sidechain contacts between the interpeptide NN, CC, and NC regions. Here, we consider a contact formed if it persists for at least 500 ps. In 10 of the 15 runs (Runs 1–10), we observe early formation of CC contacts. In the remaining five runs (Runs 11–15), we see early formation of interpeptide NC contacts. Interestingly, we never see early formation of NN contacts. These results indicate that the early stages of aggregation result from a competition between interpeptide CC and NC interactions, and CC interactions are more likely to play a dominant role. This is consistent with the proposition that the residues 92–99 play a crucial role in the fibrillization of full-length β2m (23).
TABLE 2.
Time (in nanoseconds) for first occurrence of sidechain-sidechain contacts between the interpeptide NN, CC, and NC regions
| Runs | R1 | R2 | R3 | R4 | R5 | R6 | R7 | R8 | R9 | R10 | R11 | R12 | R13 | R14 | R15 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NN | 4.1 | 3.2 | nd | nd | nd | 1.0 | 4.7 | nd | nd | nd | 3.5 | 1.5 | 3.7 | nd | 3.0 |
| CC | 3.0 | 0.8 | 2.0 | 1.7 | 2.0 | 0.5 | 1.5 | 1.0 | 2.2 | 1.7 | 2.2 | nd | 4.1 | 4.5 | nd |
| NC | 3.6 | 3.2 | nd | 3.1 | nd | nd | 1.6 | nd | nd | 1.9 | 3.5 | 1.0 | 0.3 | nd | 4.2 |
| CN | 4.8 | nd | 2.5 | nd | 2.5 | 0.7 | nd | nd | nd | 2.0 | 0.9 | 4.5 | 3.5 | 3.5 | 2.8 |
The N-terminal and C-terminal regions span H84-Q89 and I92-R97, respectively. The term nd stands for no defined interactions within 5 ns.
Interlaced β-hairpin structures are observed in two of our 200-ns MD simulations. Both simulations start from the same position coordinates with different initial velocities. An important question is whether this final intertwined structure is dictated by the starting conformation and the protonation state of the N- and C-termini. To this end, two independent 40-ns MD runs are performed at 310 K starting from a preformed antiparallel β-sheet spanning residues V93–R97 with distinct protonation states of the termini: Run D with NH2 and COO− termini and Run E with NH3+ and COOH termini. As seen in Supplementary Material Fig. S2, both simulations result in intertwined topologies with distinct hydrogen-bonding interactions within 40-ns timescale. We also perform a 60-ns MD simulation (Run F) with the NH3+ and COOH termini starting from a short antiparallel β-sheet spanning V93–R97, but with the rest of the amino acids fully extended so that the competition between two side-by-side hairpins and interlaced β-hairpins is possible. We observe that, at t = 30 ns, the C-terminal of one peptide prefers to interact with the C-terminal of the other peptide. An interlaced β-hairpin structure forms at t = 55 ns and remains until the end of the simulation (60 ns). These simulations suggest that acid pH conditions do not change the propensity for assembly into a dimeric structure with two interlaced β-hairpins. They also clearly indicate, along with our 15 short MD runs and previous studies of amyloid-forming peptides (41–44), that there are multiple aggregation routes and the dimerization pathways described in Runs A–F must be taken as indicative only.
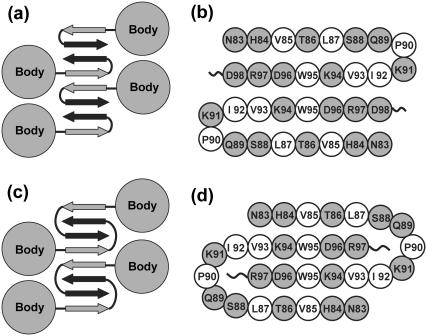
Our simulations show that two β2m(83–99) peptides have a nonnegligible probability to aggregate into a β-sheet structure. This result, in agreement with the suggestion that the C-terminal residues of full-length β2m protein are in β-sheet within the fibrils (12,45), is of interest because the dimer can act as a seed for larger oligomers and protofibrils. In this context, two fibril models for full-length β2m were recently proposed. Nussinov et al. (46) constructed a cross-β helical model based on protein docking, using NMR spectroscopy (47) and limited proteolysis data at pH ≥ 4 (48) suggesting that the N- and C-terminal β-strands (amino acids 12–18 and 91–95) do not participate in the amyloid fibril. In contrast, Eisenberg et al. recently proposed a protofibril model, where the protein core (residues 1–80) retains its native (in solution) conformation and the zipper-spine model is characterized by two hairpins, where the segment 83–89 binds to the same segment in other molecules forming the β-sheet spine of the amyloid (21). In this model, shown in Fig. 7, a and b, residues 90–91 form the β-turn, and residues 92–98 and 83–89 form the strands of β-hairpin. Interestingly, the participation of amino acids 83–99 in the β-sheet structure is supported by recent H/D exchange and limited proteolysis of full-length β2m fibrils formed at pH 2.5 at low ionic strength, demonstrating the plasticity of the morphologies and digestion patterns of fibrils as a function on the experimental conditions used (12,45).
FIGURE 7.
Two protofibril models for the full-length β2m. (a and b) Eisenberg's model with side-by-side-hairpin β-sheet dimers, and the amino acids labeled. (c and d) Our model with interlaced-hairpin-like β-sheet dimers.
Based on our simulations, we can propose an alternative model, where the hairpins are not side-by-side assembled, but rather are interlaced. Since the interlaced hairpins generated by Runs A, B, and F differ at an atomic level of detail, it is not clear which ending structure would be the most likely intertwined dimer. We therefore construct a model, shown in Fig. 7, c and d, based on three assumptions: 1), the protein core retains its in solution conformation; 2), residues 88–92 form the turn and residues 83–87 and 93–97 form the strands; and 3), the interpeptide W95-W95, K94-D96, and W95-K94 interactions (detected by our MD simulations) provide the main forces within the dimer. Our model is then modeled by maximizing the interpeptide hydrophobic interactions and hydrogen-bonding interactions (49). Interestingly, the constructed model displays a hydrophobic-network repeat V85-W95-W95-V85 along the axis of the protofibril.
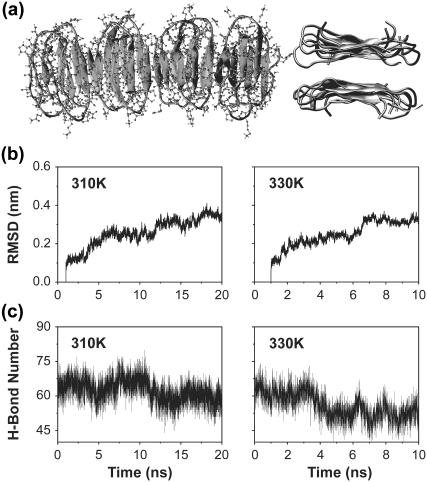
It follows that our model shares many common structural features with the model proposed by Eisenberg et al., except that the position of the turns differs and there is a β-strand swapping. Such a swapping, a consequence of minimal frustration (50), is usually seen for domains in large proteins (51–53). To determine whether our model is stable, a protofibril consisting of two layers of four units (each unit modeled by two interlaced hairpins) is subject to two MD runs at 310 K (20 ns) and 330 K (10 ns). We recognize that this timescale is too short to validate our model, but microsecond simulations are still out of reach (49). Fig. 8 a shows the initial structure for MD. The Cα RMSD from the structure at 1 ns is given in Fig. 8 b. The time evolution of the number of main chain H-bonds is shown in Fig. 8 c. Because of finite-size effects in the protofibril, we focus on the properties of the two central units of both layers. We see that the Cα RMSD increases slightly in the first five nanoseconds and then stabilizes at ∼3.0 Å, and the main chain H-bonds are rather stable at both temperatures. The increase in RMSD results from both the flexibility of each interlaced hairpin (contribution of 1.5 Å RMSD) and the twisting motion of each unit with respect to the other.
FIGURE 8.
Stability analysis of our proposed protofibril model at 310 and 330 K. (a) Two different views of the two-layer structure at t = 1 ns: (left) top view with all atoms; (right) side view along the fibril axis (side chains are not shown). (b) Time evolution of the Cα RMSD with respect to the conformation at t = 1 ns. (c) Time evolution of the total number of main-chain H-bonds. The properties in panels b and c are calculated using the two central units of both layers.
In summary, this work indicates that an intertwined dimer is a possible structure for two β2m(83–99) peptides. Whether this state is truly the most favorable or only a product of the starting conformations used remains to be determined using all-atom and coarse-grained REMD simulations (54,55). Experimental measurements, clearly needed to clarify the β-sheet packing and isotope-edited IR spectroscopy, which can differentiate between antiparallel and parallel β-sheets (56), would be helpful. However, based on insights from our dimer simulations, we propose an antiparallel arrangement of the C-terminal residues within the full-length β2m protein fibrils and the key roles of the amino acids K94, W95, and D96 in the aggregation process, leading to one of the possible organizations of β2m fibrils.
SUPPLEMENTARY MATERIAL
To view all of the supplemental files associated with this article, visit www.biophysj.org.
Supplementary Material
Acknowledgments
Simulations were performed at the Shanghai Supercomputing Center and the National High Performance Computing Center of Fudan University.
This work was supported by the National Natural Science Foundation of China (grant No. 10674029), Program for Changjiang Scholars and Innovative Research Team in University (PCSIRT), the Young Foundation, and the Senior Visiting Scholar Grant of Fudan University, and the Centre National de la Recherche Scientifique and University of Paris 7 Denis-Diderot.
Editor: John E. Straub.
References
- 1.Cohen, F. E., and J. W. Kelly. 2003. Therapeutic approaches to protein-misfolding diseases. Nature. 426:905–909. [DOI] [PubMed] [Google Scholar]
- 2.Dobson, C. M. 2003. Protein folding and misfolding. Nature. 426:884–890. [DOI] [PubMed] [Google Scholar]
- 3.Chiti, F., and C. M. Dobson. 2006. Protein misfolding, functional amyloid, and human disease. Annu. Rev. Biochem. 75:333–366. [DOI] [PubMed] [Google Scholar]
- 4.Sunde, M., L. C. Serpell, M. Bartlam, P. E. Fraser, M. B. Pepys, and C. C. Blake. 1997. Common core structure of amyloid fibrils by synchrotron x-ray diffraction. J. Mol. Biol. 273:729–739. [DOI] [PubMed] [Google Scholar]
- 5.Koch, K. M. 1992. Dialysis-related amyloidosis. Kidney Int. 41:1416–1429. [DOI] [PubMed] [Google Scholar]
- 6.Miyata, T., M. Jadoul, K. Kurokawa, and C. Van Ypersele de Strihou. 1998. β2-Microglobulin in renal disease. J. Am. Soc. Nephrol. 9:1723–1735. [DOI] [PubMed] [Google Scholar]
- 7.Floege, J., and G. Ehlerding. 1996. β2-Microglobulin-associated amyloidosis. Nephron. 72:9–36. [DOI] [PubMed] [Google Scholar]
- 8.Gejyo, F., N. Homma, Y. Suzuki, and M. Arakawa. 1986. Serum levels of β2-microglobulin as a new form of amyloid protein in patients undergoing long-term hemodialysis. N. Engl. J. Med. 314:585–586. [DOI] [PubMed] [Google Scholar]
- 9.McParland, V. J., N. M. Kad, A. P. Kalverda, A. Brown, P. Kirwin-Jones, M. G. Hunter, M. Sunde, and S. E. Radford. 2000. Partially unfolded states of β2-microglobulin and amyloid formation in vitro. Biochemistry. 39:8735–8746. [DOI] [PubMed] [Google Scholar]
- 10.Kad, N. M., N. H. Thomson, D. P. Smith, D. A. M. Smith, and S. E. Radford. 2001. β2-Microglobulin and its deamidated variant, N17D form amyloid fibrils with a range of morphologies in vitro. J. Mol. Biol. 313:559–571. [DOI] [PubMed] [Google Scholar]
- 11.Smith, D. P., and S. E. Radford. 2001. Role of the single disulphide bond of β2-microglobulin in amyloidosis in vitro. Protein Sci. 10:1775–1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Myers, S. L., N. H. Thomson, S. E. Radford, and A. E. Ashcroft. 2006. Investigating the structural properties of amyloid-like fibrils formed in vitro from β2-microglobulin using limited proteolysis and electrospray ionization mass spectrometry. Rapid Commun. Mass Spectrom. 20:1628–1636. [DOI] [PubMed] [Google Scholar]
- 13.Armen, R. S., and V. Daggett. 2005. Characterization of two distinct β2-microglobulin unfolding intermediates that may lead to amyloid fibrils of different morphology. Biochemistry. 44:16098–16107. [DOI] [PubMed] [Google Scholar]
- 14.Chen, Y., and N. V. Dokholyan. 2005. A single disulfide bond differentiates aggregation pathways of β2-microglobulin. J. Mol. Biol. 354:473–482. [DOI] [PubMed] [Google Scholar]
- 15.Nishino, M., Y. Sugita, T. Yoda, and Y. Okamoto. 2005. Structures of a peptide fragment of β2-microglobulin studied by replica-exchange molecular dynamics simulations—towards the understanding of the mechanism of amyloid formation. FEBS Lett. 579:5425–5429. [DOI] [PubMed] [Google Scholar]
- 16.Lei, H., C. Wu, Z. Wang, and Y. Duan. 2006. Molecular dynamics simulations and free energy analyses on the dimer formation of an amyloidogenic heptapeptide from human β2-microglobulin: implication to the protofibril structure. J. Mol. Biol. 356:1049–1063. [DOI] [PubMed] [Google Scholar]
- 17.Kozhukh, G. V., Y. Hagihara, T. Kawakami, H. Hasegawa, K. Naiki, and Y. Goto. 2002. Investigation of a peptide responsible for amyloid fibril formation of β2-microglobulin by Achromobacter protease i. J. Biol. Chem. 277:1310–1315. [DOI] [PubMed] [Google Scholar]
- 18.Jones, S., J. Manning, N. M. Kad, and S. E. Radford. 2003. Amyloid-forming peptides from β2-microglobulin: insights into the mechanism of fibril formation in vitro. J. Mol. Biol. 325:249–257. [DOI] [PubMed] [Google Scholar]
- 19.Lu, M., H. Hiramatsu, Y. Goto, and T. Kitagawa. 2006. Structure of interacting segments in the growing amyloid fibril of β2-microglobulin probed with IR spectroscopy. J. Mol. Biol. 362:355–364. [DOI] [PubMed] [Google Scholar]
- 20.Ivanova, M. I., M. Gingery, L. J. Whitson, and D. Eisenberg. 2003. Role of the C-terminal 28 residues of β2-microglobulin in amyloid fibril formation. Biochemistry. 42:13536–13540. [DOI] [PubMed] [Google Scholar]
- 21.Ivanova, M. I., M. R. Sawaya, M. Gingery, A. Attinger, and D. Eisenberg. 2004. An amyloid-forming segment of β2-microglobulin suggests a molecular model for the fibril. Proc. Natl. Acad. Sci. USA. 101:10584–10589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ivanova, M. I., M. J. Thompson, and D. Eisenberg. 2006. A systematic screen of β2-microglobulin and insulin for amyloid-like segments. Proc. Natl. Acad. Sci. USA. 103:4079–4082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stoppini, M., V. Bellotti, P. Mangione, G. Merlini, and G. Ferri. 1997. Use of anti-(β2-microglobulin) mAb to study formation of amyloid fibrils. Eur. J. Biochem. 249:21–26. [DOI] [PubMed] [Google Scholar]
- 24.Nelson, R., and D. Eisenberg. 2006. Recent atomic models of amyloid fibril structure. Curr. Opin. Struct. Biol. 16:260–265. [DOI] [PubMed] [Google Scholar]
- 25.Berg, J. M., L. Stryer, and J. L. Tymoczko. 2002. Biochemistry, 5th Ed. W. H. Freeman, New York.
- 26.Berendsen, H. J. C. 1981. Interaction Models for Water in Relation to Protein Hydration, Intermolecular Forces. Reidel, Dordrecht, The Netherlands.
- 27.Lindahl, E., B. Hess, and D. Van der Spoel. 2001. GROMACS 3.0: a package for molecular simulation and trajectory analysis. J. Mol. Mod. 7:306–317. [Google Scholar]
- 28.Van Gunsteren, W. F., S. R. Billeter, A. A. Eising, P. H. Hunenberger, P. Kruger, A. E. Mark, W. R. P. Scott, and I. G. Tironi. 1996. Biomolecular Simulation: The GROMOS96 Manual and User Guide. Vdf Hochschulverland, ETH, Zurich, Switzerland.
- 29.Berendsen, H. J. C., J. P. M. Postma, A. Di Nola, and J. R. Haak. 1984. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 81:3684–3690. [Google Scholar]
- 30.Kabsch, W., and C. Sander. 1983. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 22:2577–2637. [DOI] [PubMed] [Google Scholar]
- 31.Wei, G. H., and J. E. Shea. 2006. Effects of solvent on the structure of the Alzheimer amyloid-β(25–35) peptide. Biophys. J. 91:1638–1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baumketner, A., and J. E. Shea. 2006. Folding landscapes of the Alzheimer amyloid-β(12–28) peptide. J. Mol. Biol. 362:567–579. [DOI] [PubMed] [Google Scholar]
- 33.Hashioka, S., A. Monji, T. Ueda, S. Kanba, and H. Nakanishi. 2005. Amyloid-β fibril formation is not necessarily required for microglial activation by the peptides. Neurochem. Int. 47:369–376. [DOI] [PubMed] [Google Scholar]
- 34.Yoda, T., Y. Sugita, and Y. Okamoto. 2004. Comparisons of force fields for proteins by generalized-ensemble simulations. Chem. Phys. Lett. 386:460–467. [Google Scholar]
- 35.Baumketner, A., and J. E. Shea. 2007. The structure of the Alzheimer amyloid-β (10–35) peptide probed through replica-exchange molecular dynamics simulations in explicit solvent. J. Mol. Biol. 366:275–285. [DOI] [PubMed] [Google Scholar]
- 36.Wu, C., H. Lei, and Y. Duan. 2005. Elongation of ordered peptide aggregate of an amyloidogenic hexapeptide NFGAIL observed in molecular dynamics simulations with explicit solvent. J. Am. Chem. Soc. 127:13530–13537. [DOI] [PubMed] [Google Scholar]
- 37.Platt, G. W., V. J. McParland, A. P. Kalverda, S. W. Homans, and S. E. Radford. 2005. Dynamics in the unfolded state of β2-microglobulin studied by NMR. J. Mol. Biol. 346:279–294. [DOI] [PubMed] [Google Scholar]
- 38.Tjernberg, L., W. Hosia, N. Bark, J. Thyberg, and J. Johansson. 2002. Charge attraction and β propensity are necessary for amyloid fibril formation from tetrapeptides. J. Biol. Chem. 277:43243–43246. [DOI] [PubMed] [Google Scholar]
- 39.Chiti, F., M. Stefani, N. Taddei, G. Ramponi, and C. M. Dobson. 2003. Rationalization of the effects of mutations on peptide and protein aggregation rates. Nature. 424:805–808. [DOI] [PubMed] [Google Scholar]
- 40.Yoon, S., and W. J. Welsh. 2005. Rapid assessment of contact-dependent secondary structure propensity: relevance to amyloidogenic sequences. Proteins. 60:110–117. [DOI] [PubMed] [Google Scholar]
- 41.Santini, S., G. H. Wei, N. Mousseau, and P. Derreumaux. 2004. Pathway complexity of Alzheimer's β-amyloid aβ(16–22) peptide assembly. Structure. 12:1245–1255. [DOI] [PubMed] [Google Scholar]
- 42.Santini, S., N. Mousseau, and P. Derreumaux. 2004. In silico assembly of Alzheimer's aβ(16–22) peptide into β-sheets. J. Am. Chem. Soc. 37:11509–11516. [DOI] [PubMed] [Google Scholar]
- 43.Wei, G. H., N. Mousseau, and P. Derreumaux. 2004. Sampling the self-assembly pathways of KFFE hexamers. Biophys. J. 87:3648–3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Melquiond, A., J. C. Gelly, N. Mousseau, and P. Derreumaux. 2007. Probing amyloid fibril formation of the NFGAIL peptide by computer simulations. J. Chem. Phys. 126:065101–065107. [DOI] [PubMed] [Google Scholar]
- 45.Yamaguchi, K., H. Katou, M. Hoshino, K. Hasegawa, H. Naiki, and Y. Goto. 2004. Core and heterogeneity of β2-microglobulin amyloid fibrils as revealed by H/D exchange. J. Mol. Biol. 338:559–571. [DOI] [PubMed] [Google Scholar]
- 46.Benyamini, H., K. Gunasekaran, H. Wolfson, and R. Nussinov. 2003. β2-Microglobulin amyloidosis: insights from conservation analysis and fibril modeling by protein docking techniques. J. Mol. Biol. 330:159–174. [DOI] [PubMed] [Google Scholar]
- 47.McParland, V. J., A. P. Kalverda, S. W. Homans, and S. E. Radford. 2002. Structural properties of an amyloid precursor of β2-microglobulin. Nat. Struct. Biol. 9:326–331. [DOI] [PubMed] [Google Scholar]
- 48.Monti, M., S. Principe, S. Giorgetti, P. Mangione, G. Merlini, A. Clark, V. Bellotti, A. Amoresano, and P. Pucci. 2002. Topological investigation of amyloid fibrils obtained from β2-microglobulin. Protein Sci. 11:2362–2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ma, B., and R. Nussinov. 2006. Simulations as analytical tools to understand protein aggregation and predict amyloid conformation. Curr. Opin. Chem. Biol. 10:445–452. [DOI] [PubMed] [Google Scholar]
- 50.Yang, S., S. S. Cho, Y. Levy, M. S. Cheung, H. Levine, P. G. Wolynes, and J. N. Onuchic. 2004. Domain swapping is a consequence of minimal frustration. Proc. Natl. Acad. Sci. USA. 101:13786–13791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Huntington, J. A., N. S. Pannu, B. Hazes, R. J. Read, D. A. Lomas, and R. W. Carrell. 1999. A 2.6 Å structure of a serpin polymer and implications for conformational disease. J. Mol. Biol. 293:449–455. [DOI] [PubMed] [Google Scholar]
- 52.Janowski, R., M. Kozak, E. Jankowska, Z. Grzonka, A. Grubb, M. Abrahamson, and M. Jaskolski. 2001. Human cystatin C, an amyloidogenic protein, dimerizes through three-dimensional domain swapping. Nat. Struct. Biol. 8:316–320. [DOI] [PubMed] [Google Scholar]
- 53.Eakin, C. M., F. J. Attenello, C. J. Morgan, and A. D. Miranker. 2004. Oligomeric assembly of native-like precursors precedes amyloid formation by β2-microglobulin. Biochemistry. 43:7808–7815. [DOI] [PubMed] [Google Scholar]
- 54.Derreumaux, P., and N. Mousseau. 2007. Coarse-grained protein molecular dynamics simulations. J. Chem. Phys. 126:025101–025106. [DOI] [PubMed] [Google Scholar]
- 55.Wei, G. H., N. Mousseau, and P. Derreumaux. 2007. Computational simulations of the early steps of protein aggregation. Prion. 1:1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Petty, S. A., and S. M. Decatur. 2005. Intersheet rearrangement of polypeptides during nucleation of β-sheet aggregates. Proc. Natl. Acad. Sci. USA. 102:14272–14277. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.