Abstract
Maturation of Physarum mitochondrial RNA involves the highly specific insertion of nonencoded nucleotides at multiple locations. To investigate the mechanism(s) by which this occurs, we previously developed an isolated mitochondrial system in which run-on transcripts are accurately and efficiently edited by nucleotide insertion. Here we show that under limiting concentrations of exogenous nucleotides the mitochondrial RNA polymerases stall, generating a population of nascent RNAs that can be extended upon addition of limiting nucleotide. Several of these RNA species have been characterized and were found to be fully edited, indicating that nascent RNA is a substrate for nucleotide insertion in isolated Physarum mitochondria. Remarkably, these RNAs are edited at positions located within 14–22 nucleotides of the polymerase active site, suggesting that insertional editing may be physically or functionally associated with transcription. The absence of unedited RNA in these experiments indicates that large tracts of RNA downstream of editing sites are not required for nucleotide addition, and argues that insertional editing in Physarum occurs with a 5′ to 3′ polarity. These data also provide strong evidence that insertional editing in Physarum is mechanistically distinct from editing in kinetoplastid systems.
Keywords: RNA editing, nucleotide insertion, transcription, mitochondrial RNA polymerase, Physarum polycephalum
The term RNA editing is used to describe two distinct types of alterations to RNA: base substitution (or conversion) and nucleotide insertion/deletion (reviewed in ref. 1). The mitochondrial RNAs of the slime mold Physarum polycephalum are unique in that they are processed by both types of editing (2), each of which is extremely efficient in vivo (2, 3). Most editing events in Physarum involve the insertion of single cytidine (C) residues at defined sites, although the specific addition of each of the four nucleotides as single or dinucleotide insertions into mRNA, tRNA, and rRNA has been described (2, 4–6). In addition, the cytochrome c oxidase subunit one (coI) transcript is processed by four apparent C to U changes, as well as the insertion of 59 single Cs, a single uridine (U), and the dinucleotides UA, CU, and GU (2, 7). However, despite the large number of editing sites that have been characterized, the cis-acting signals and cellular machinery responsible for both the specificity and diversity of the observed editing events in Physarum mitochondrial RNAs have yet to be identified.
Comparison of the patterns of nucleotide addition in Physarum with that of other well-characterized insertional editing systems suggests that a unique mechanism is likely to be employed. For example, RNA editing in paramyxoviruses and Ebola virus occurs through the cotranscriptional insertion of one or more nucleotides at a homopolymer tract within the template (8–13). A strictly analogous mechanism is unlikely in Physarum, as most editing sites are flanked by nucleotides other than that inserted (2, 4, 6). Insertional editing in Physarum is also likely to be mechanistically distinct from the uridine addition and deletion that occurs in the kinetoplastid protozoa (1, 14). Editing in kinetoplasts is a posttranscriptional process mediated by guide RNAs that direct endonuclease cleavage at individual editing sites and specify the number of residues added or deleted at that site (15–17). Thus far, there is no evidence for nucleotide deletion in Physarum mitochondrial RNAs, nor have attempts to locate guide-like RNAs been successful (ref. 5 and unpublished results). In addition, both the number and spacing of nucleotide insertions differ between the two systems, with editing of Physarum RNAs usually involving single, widely spaced insertions (2, 4, 6) rather than intensive editing “blocks” (1). Finally, nucleotide addition in Physarum is complicated by the ability of the editing machinery to insert each of the four nucleotides in a site-specific manner (2, 6).
The lack of parallels with the known insertional editing systems has led us to investigate the nature of editing substrates in Physarum to determine whether editing in this organism occurs through a cotranscriptional or posttranscriptional process. To address this issue experimentally, we have used a recently developed isolated mitochondrial system in which labeled RNAs can be synthesized and processed under defined conditions (7). RNAs made in this in vitro system are accurately and efficiently processed by nucleotide insertion, but are not edited by C to U changes (7). Here we show that by limiting exogenously supplied nucleotides in the mitochondrial transcription reactions, labeled RNAs derived from stalled ternary complexes can be isolated. Analysis of these RNAs shows that nascent transcripts are an editing substrate, and that the Physarum insertional editing activity can function very close to the site of RNA synthesis.
MATERIALS AND METHODS
All experimental procedures were performed as previously described (7), except as noted.
RNA Synthesis and S1 Nuclease Protection Experiments.
To generate radiolabeled RNAs, the crude mitochondrial preparation (1.45 mg/ml) was preincubated for 3 min in transcription buffer [20 mM Tris⋅HCl, pH 7.5 (at 25°C)/20 mM MgCl2/10 mM KCl/2 mM DTT] at 35°C to deplete endogenous nucleoside triphosphate (NTP) pools prior to use. Pulse-labeling conditions: 175 μM unlabeled NTPs (nonlimiting), 200 nM [α-32P]NTP (limiting); pulse–chase conditions: 100 μM of the specified NTP was added after a 3 min labeling reaction and incubated as specified. S1 nuclease digestions were carried out as previously described in a buffer containing 0.28 M NaCl, 0.05 M sodium acetate (pH 4.5), 4.5 mM ZnSO4, except that protected mitochondrial RNAs were reprecipitated with 1 μg of single-stranded DNA (ssDNA), annealed at 37°C for 2 hr, and subjected to a second round of S1 nuclease digestion to ensure complete digestion prior to separation on 6% denaturing polyacrylamide gels. The S1 nuclease protection experiment in Fig. 2 was performed in a modified buffer (0.75 M NaCl/0.05 M sodium acetate, pH 4.5/4.5 mM ZnSO4) to minimize breathing of the RNA–DNA hybrids during digestion. Under these conditions, three rounds of S1 nuclease digestion were required to achieve efficient cleavage of both control transcripts and mitochondrial RNAs at the GU insertion site when hybridized to the unedited probe. Control RNA transcripts (nt 85–1778) of the edited coI mRNA sequence (2) were synthesized using T7 RNA polymerase (Ambion MAXIscript kit).
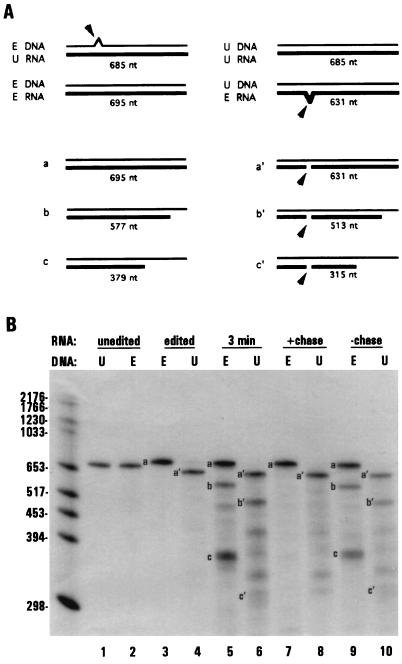
Figure 2.
RNAs associated with stalled transcription complexes are edited by dinucleotide insertion. (A) Schematic diagram of dinucleotide insertion assay. Arrows indicate dinucleotide bulges present in hybrids between edited (E) and unedited (U) molecules that are preferentially cleaved by S1 nuclease. Single nucleotide bulges are not cleaved efficiently and are therefore not depicted. Also shown are schematic diagrams of the protection patterns and observed sizes of labeled mitochondrial RNAs produced under limiting GTP conditions (thick lines). (B) Unlabeled ssDNA having either edited (E, probe 1, lanes 2, 3, 5, 7, and 9) or the corresponding unedited (U, probe 2, lanes 1, 4, 6, 8, and 10) coI sequence were used in S1 nuclease protection of labeled RNAs. Lanes 1–4, S1 protection of unedited (lanes 1 and 2) and edited (lanes 3 and 4) coI control transcripts. Lanes 5–10, RNAs synthesized in isolated mitochondria. Lanes 5 and 6, 3-min pulse labeling, limiting GTP; lanes 7 and 8, 3-min pulse labeling, limiting GTP, followed by a 12 min chase with 100 μM GTP; lanes 9 and 10, RNAs synthesized during a 15-min pulse labeling, limiting GTP. S1 protected samples were separated on a 4% denaturing polyacrylamide gel. The 5′ cleavage product resulting from S1 protection with probe 2 (U) was run off of the gel.
Probes.
ssDNAs used in S1 nuclease protection experiments correspond to the following regions of the coI sequence (2): probes 1 (edited) and 2 (unedited), nt 1084–1778; probes 3a (unedited) and 3b (edited), nt 443–654; probes 4a (unedited) and 4b (edited), nt 443–807. Probe 3c (nt 443–906) and 4c (nt 443–807) have the edited sequence at all insertion sites, but unedited sequence at the sites of C to U substitutions located at nucleotide positions 734, 736, and 737 to allow efficient protection of RNAs synthesized in isolated mitochondria, which are not edited by C to U changes (7).
Separation and Analysis of RNase T1 Oligonucleotides.
S1-protected RNAs for use in RNase T1 assays were purified on 5% denaturing gels; for fingerprint analyses, mitochondrial RNAs were reisolated from a 4% denaturing gel to reduce nonspecific background. RNase T1 oligonucleotides were separated in one dimension on 20% polyacrylamide/7 M urea/50 mM Tris/50 mM boric acid/1 mM EDTA, pH 8.3 (TBE) gels or in two dimensions by RNA fingerprinting (7). For secondary analyses, isolated RNase T1 oligonucleotides were digested to ribonucleoside 3′ monophosphates upon the addition of a ribonuclease mixture containing RNase T2, RNase T1, and RNase A, and separated by two-dimensional chromatography as described (7).
RESULTS AND DISCUSSION
Polymerase Stalling Under Limiting NTP Concentrations.
To analyze the editing patterns of individual nascent species, we first isolated RNAs from transcription complexes that were stalled at specific sites within the coI gene. When the concentration of a single nucleotide is limited during run-on transcription, discrete populations of RNAs are produced due to polymerase stalling at various points along the mitochondrial genome (Fig. 1). Individual species can be detected and isolated using S1 nuclease protection with specific ssDNA probes. We have previously shown that under the conditions used in these experiments both edited and unedited control transcripts are fully protected when isolated with the edited probe (7). Protection of labeled mitochondrial RNAs synthesized in the presence of each of the four limiting nucleotides during a 3-min pulse labeling is shown in Fig. 1B (lanes 2–5). In addition to an RNA that is the length of the cytochrome c oxidase subunit 1 (coI) probe, distinct shorter RNA species were also detected with each limiting nucleotide. The shorter species were not produced as the result of nonspecific S1 nuclease cleavage, since only a single RNA species was protected when a control transcript was hybridized to the same probe (lane 1). Oligodeoxynucleotide-directed RNase H analysis of a large number of these shorter RNAs has confirmed that in each case the observed heterogeneity was due to differences at the 3′ ends of the RNA species (data not shown).
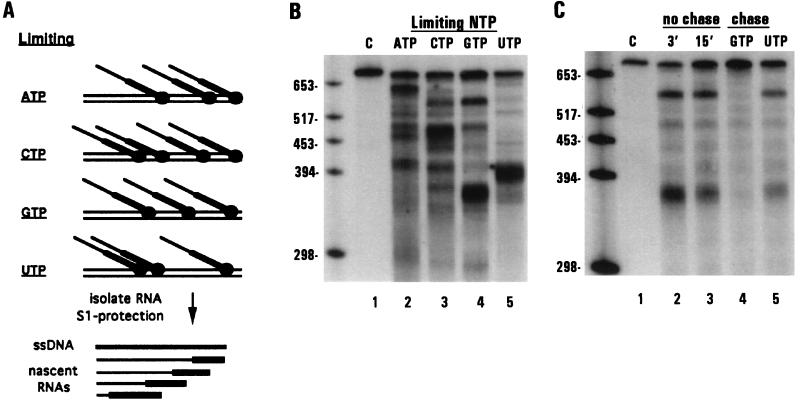
Figure 1.
S1 nuclease protection of shorter RNAs associated with stalled transcription complexes. (A) Schematic diagram of RNAs synthesized under limiting nucleotide concentrations in isolated mitochondria and S1 nuclease protection of the resulting RNA species. (B and C) S1 nuclease protection of labeled RNAs with unlabeled ssDNA corresponding to edited coI sequence (probe 1). (B) Lane 1, edited coI control transcript; lanes 2–5, RNAs synthesized in isolated mitochondria during a 3-min pulse labeling in the presence of limiting (radiolabeled) ATP, CTP, GTP, or UTP. (C) Lane 1, edited coI control transcript; lanes 2–5, RNAs synthesized in isolated mitochondria. Lanes: 2, 3-min pulse labeling, limiting GTP; 3, 15-min pulse labeling, limiting GTP; 4, 3-min pulse labeling, limiting GTP, followed by a 12 min chase with 100 μM GTP; 5, 3-min pulse labeling, limiting GTP, followed by a 12 min chase with 100 μM UTP.
To verify that the shorter transcripts were associated with stalled transcription complexes, as opposed to products of premature termination, we examined whether these RNAs could be extended. In one such experiment, mitochondrial RNAs were pulse labeled for 3 min under limiting GTP conditions and an aliquot was removed to confirm the production of the shorter RNA species (Fig. 1C, lane 2). The remaining sample was split into thirds and incubated in either the presence or absence of additional NTPs as specified (lanes 3–5). When the mitochondria were incubated with additional GTP, the shorter species were extended into longer RNAs (lane 4). Extension of these RNAs has been confirmed by a combination of RNase H and RNase T1 fingerprinting experiments (data not shown). In contrast, the shorter RNAs were neither extended nor degraded during a 12 min incubation without the addition of limiting nucleotide (lane 3) or when chased with the nonlimiting nucleotide UTP (lane 5). Similar results were obtained in separate experiments when other nucleotides were limiting (data not shown). Thus, by limiting the supply of exogenous nucleotides provided to isolated Physarum mitochondria, specific nascent RNA species were produced that were associated with stalled transcription complexes. Interestingly, stalling was observed at only a subset of the potential sites for each nucleotide, suggesting that sequence context also plays a role in polymerase stalling in isolated Physarum mitochondria.
Processing of Nascent RNAs by GU Insertion.
To determine if these nascent RNAs were edited at the GU dinucleotide insertion site within the coI mRNA, we used a previously described S1 nuclease assay (7) (Fig. 2A). This assay takes advantage of the ability of S1 nuclease to cleave preferentially at dinucleotide bulges present in hybrids formed between edited and unedited molecules (Fig. 2B, lanes 1–4). When RNA synthesized in isolated mitochondria under limiting GTP concentrations was protected with the edited probe (Fig. 2B, lanes 5 and 9), both full-length (species a) and shorter (species b and c) RNAs were produced. However, when the same mitochondrial RNA sample was protected with the unedited probe, species a, b, and c were absent, whereas new fragments (a′, b′, c′) were observed whose sizes and RNase H digestion patterns were consistent with cleavage at the GU insertion site (lanes 6 and 10 and data not shown). Extension of the shorter RNAs upon incubation with additional GTP confirmed that they were associated with stalled polymerases (lane 7). As expected, the extended RNA species were also efficiently edited at the dinucleotide insertion site (lane 8). These data demonstrate that nascent RNAs are a substrate for editing by dinucleotide insertion.
Editing of Nascent RNAs Near the Site of Transcription.
To investigate further the temporal relationship between insertional editing and transcription in Physarum, we next asked how close editing could be detected to the 3′ end of nascent RNAs. If regions of the RNA near the site of transcription were unedited, this would provide evidence that the editing apparatus functions independently of transcription. Conversely, the presence of added nucleotides close to the end of the growing RNA chain would suggest that editing is associated with transcription. We have used RNA fingerprint analyses to examine editing of several nascent RNA species at insertion sites that fall within 25 nt of their respective 3′ ends. A representative analysis is shown in Fig. 3. In this experiment, RNA was synthesized in isolated mitochondria in the presence of limiting (radiolabeled) ATP and was protected from S1 nuclease digestion with a coI-specific probe. The 3′ end of one protected species was mapped to RNase T1 oligonucleotide q, where a run of five adenosine residues is encoded in the DNA (bracketed in Fig. 3). Because polymerases in this region may have stalled anywhere within the run of adenosine residues, the 3′ ends of these RNAs fell between 18 and 22 nt from the C insertion site present in RNase T1 oligonucleotide p, and only 3–8 nt from the C insertion site within oligonucleotide q. Pulse–chase studies similar to those shown in Figs. 1 and 2 have verified that this RNA was extended upon the addition of ATP, but not when other nonlimiting nucleotides were provided to isolated mitochondria (data not shown).
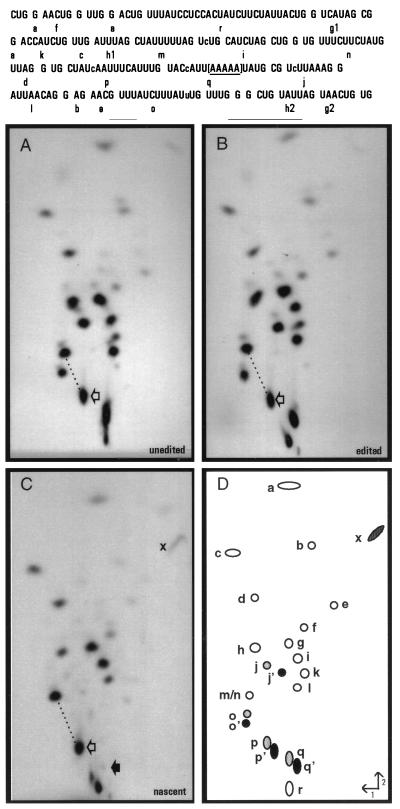
Figure 3.
RNase T1 fingerprint analyses. (Upper) Sequence of the region of interest with individual T1 oligonucleotides shown. Note that as [α-32P]ATP was used to label each RNA, only the indicated RNase T1 oligonucleotides derived from this region of the coI mRNA are visible. The bracketed area represents the region in which the transcription complexes stall under limiting ATP conditions in isolated mitochondria. Lowercase letters within the sequence indicate sites of nucleotide insertion. (Lower) [α-32P]ATP-labeled RNAs were gel purified after S1 protection with coI-specific probes (A–C, probes 3 a–c, respectively), digested with ribonuclease T1 and the resulting oligonucleotides separated in two dimensions. Fingerprints derived from (A) unedited coI control transcript, (B) edited coI control transcript, (C) nascent RNA synthesized in isolated mitochondria. RNase T1 oligonucleotides shown schematically in D were identified by secondary analysis and differential labeling of specific RNase T1 oligonucleotides in control fingerprints. (○), oligonucleotides not affected by editing; ( ), position of oligonucleotides without inserted nucleotides; (•), oligonucleotides with inserted nucleotides. For oligonucleotides overlapping sites of nucleotide insertion, each apostrophe designates the presence of an added nucleotide. Open arrows indicate oligonucleotides p and p′; the solid arrow in C indicates the position where oligonucleotide q/q′ would be located if present. When oligonucleotide p′ is processed by C insertion, its mobility is reduced in both dimensions, resulting in a shift in the fingerprint pattern downward and to the right with respect to oligonucleotides m/n. The difference in mobility of oligonucleotide p/p′ in unedited and edited RNAs relative to oligonucleotides m/n is highlighted by the dotted line. x indicates the terminal fragment in nascent RNA that is derived from the 5′ end of oligonucleotide q. Labeled mitochondrial RNAs were synthesized in the presence of 150 μM CTP, GTP, and UTP and 200 nM [α-32P]ATP.
The location of the 3′ end of the nascent mitochondrial RNA was confirmed by comparison of its fingerprint pattern with that of unedited and edited control RNAs extending approximately 70 nt further than the mitochondrial species (Fig. 3 A and B) and that of a HgaI control transcript ending within the run of A residues located in oligonucleotide q (data not shown). As expected, the nascent RNA species lacked all fragments downstream of oligonucleotide q (Fig. 3C) and its terminal RNase T1 oligonucleotide (x, truncated within q) had a mobility similar to that found in the fingerprint of the S1-protected HgaI control transcript. This result strongly suggests that the mitochondrial species was generated by polymerase stalling within oligonucleotide q.
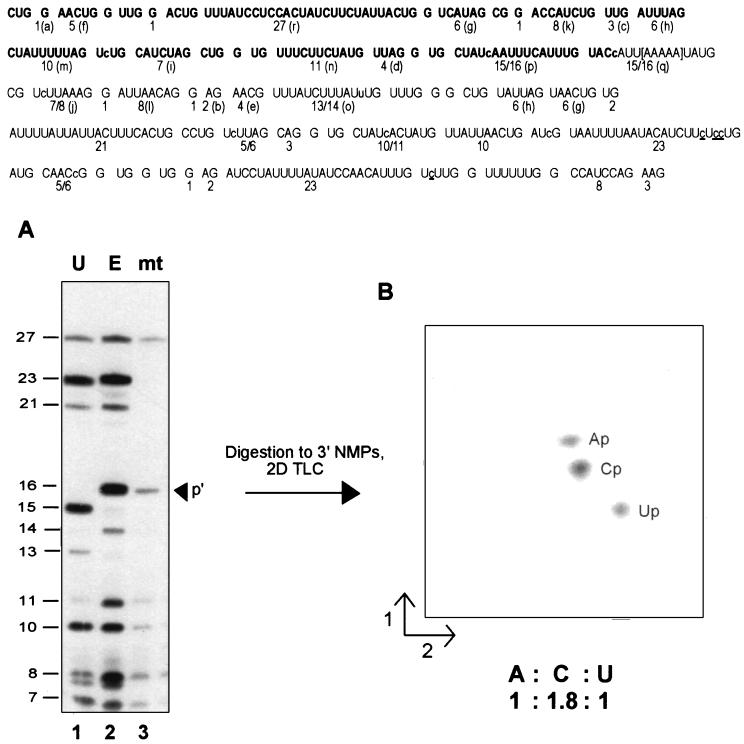
The C insertion site present just 18–22 nt from the 3′ end of this nascent RNA was accurately edited (Fig. 3C), since the relative mobility of RNase T1 oligonucleotide p is identical to that observed in the edited control fingerprint. Separation of the RNase T1 digestion products derived from this RNA species and from control transcripts extending approximately 220 nt further than the mitochondrial species on a denaturing 20% acrylamide gel (Fig. 4A) confirmed that the nascent RNA species was indeed processed by the insertion of a single nucleotide within oligonucleotide p, as an RNase T1 oligonucleotide with a size identical to the edited control (16 nt) was observed (Fig. 4A, lane 3). Editing at this site was highly efficient, because no oligonucleotide corresponding to the unedited species was observed by either method. To verify that the editing site present in oligonucleotide p′ was processed by the addition of a C residue, this fragment was isolated, digested to mononucleotides, and the resulting 3′ NMPs were separated via two-dimensional thin-layer chromatography (Fig. 4B). Because the RNA was labeled with [α-32P]ATP, digestion of the correctly edited species (CUAUcAAUUUCAUUUG) should result in transfer of label to Ap, Cp, and Up in a 1:2:1 ratio, whereas an unedited fragment (CUAUAAUUUCAUUUG) would have labeled Ap, Cp, and Up in a 1:1:2 ratio. Quantitation of the 3′ NMP digestion products in Fig. 4B indicated that the ratio of Ap:Cp:Up was 1:1.8:1, confirming that the C insertion site present within RNase T1 oligonucleotide p′ was accurately edited. Other nascent RNA species, including RNAs having 3′ ends falling within 14–28 nt of other C insertion sites and the GU and UA dinucleotide insertion sites, were also edited at all sites analyzed (data not shown). Thus, we conclude that the insertional editing apparatus can efficiently process an insertion site within 14–22 nt of the 3′ end of an RNA associated with a stalled transcription complex.
Figure 4.
Denaturing gel separation and secondary analyses. (Upper) Sequence of the RNase T1 oligonucleotides present in the S1 nuclease-protected region. The size of each [α-32P]ATP-labeled fragment is indicated; for ease of comparison, the letter assignments from Fig. 3 are shown in parentheses. The region in which the transcription complexes stall under limiting ATP conditions in isolated mitochondria is shown in brackets. Note that while the control transcripts on this gel extend 152 nt beyond those shown in the previous figure, the nascent mitochondrial RNA has the same 3′ end as that shown in Fig. 3. RNase T1 oligonucleotides visible in the mitochondrial RNA fingerprint (Fig. 3C) are indicated in boldface. Nucleotide insertion sites are shown in lowercase letters within the sequence; sites of C to U changes are underlined. (Lower) (A) [α-32P]ATP-labeled RNAs were gel purified after S1 nuclease protection with coI-specific probes (probes 4 a–c, respectively), digested with ribonuclease T1, and the resulting oligonucleotides separated on a denaturing 20% polyacrylamide gel. Oligonucleotide sizes are indicated at left. Lanes: 1, unedited coI control transcript; 2, edited coI control transcript; 3, nascent RNA synthesized in isolated mitochondria in the presence of 150 μM CTP, GTP, and UTP and 200 nM [α-32P]ATP. The arrowhead indicates the position of RNase T1 oligonucleotide p′ in the mitochondrial RNA sample. (B) RNase T1 oligonucleotide p′ from the sample in lane 3 was eluted from the gel, digested to mononucleotides, and the resulting 3′ NMPs separated via two-dimensional thin-layer chromatography as described.
Implications for the Mechanism of Insertional Editing in Physarum.
These studies provide a first step in understanding the substrates and timing of insertional editing in Physarum. We have demonstrated that nascent RNA is a substrate for insertional editing, and that nonencoded nucleotides can be added near the site of transcription. These data exclude the involvement of RNA sequences located more than 22 nt downstream in editing of these insertion sites. Despite analysis of a number of RNA species generated with various limiting nucleotides, we have not been able to definitively address whether nascent RNAs are edited closer than 14 nt from their 3′ ends. In the experiment shown in Fig. 3, for example, the sequence of oligonucleotide x precluded the use of nearest neighbor analysis to determine whether a C was inserted at the site 3–8 nt from the 3′ end of this nascent RNA. However, it should be emphasized that none of our data exclude the possibility that insertional editing may occur at sites much closer to, or even at, the site of transcription.
By the same token, because these experiments were carried out under conditions of polymerase stalling, we cannot rule out the possibility that the editing activity normally functions at a distance from the transcriptional apparatus and is merely “catching up” with stalled polymerases in these experiments. Nevertheless, our data clearly show that the Physarum editing apparatus is capable of functioning very close to the site of RNA synthesis. This finding, coupled with the fact that unedited RNAs are not detected under nonlimiting nucleotide concentrations in vivo (2, 3) or in vitro (7), raises the interesting possibility that editing and transcription may be mechanistically or physically associated with one another. Cotranscriptional editing in Ebola and paramyxoviruses is catalyzed by the viral polymerase, but in these cases nonencoded nucleotides are added only occasionally at a single “slippery” site within the template (12, 18, 19). In contrast, editing in Physarum involves the highly efficient insertion of different nucleotides in a variety of sequence contexts within most mitochondrial RNAs. Thus, even if the same underlying mechanism were used in Physarum mitochondria, both the signals for nucleotide addition and the catalytic machinery are likely to differ significantly between the two systems.
Finally, the data presented here allow clear distinctions to be made between the mechanisms of editing in Physarum and kinetoplastid protozoa (14). First, whereas insertional editing in kinetoplasts occurs in a 3′ to 5′ direction (20–22), the absence of nascent RNAs that are unedited is indicative of an overall 5′ to 3′ polarity in Physarum. Previous studies using reverse transcriptase–PCR products derived from partially edited Physarum mitochondrial RNAs showed no overall polarity of editing (described in refs. 5 and 23). However, our data suggest that these rare, partially edited species are likely to be dead-end products, rather than true editing intermediates. Second, insertional editing has been shown to occur posttranscriptionally in kinetoplasts (15, 16); in contrast, we have shown here that Physarum editing can occur at sites located within 14–22 nt of the 3′ end of the growing RNA chain. Finally, this juxtaposition of transcription and editing also suggests that nucleotide insertion in Physarum is not mediated by kinetoplastid-like guide RNAs (17). Because the editing substrate is still associated with the transcriptional machinery, very little of the RNA downstream of editing sites would be accessible for binding by guide-like RNAs (24), consistent with the fact that analogous RNAs have not been detected in Physarum. Taken together, these data indicate that insertional editing in Physarum occurs via a unique mechanism.
Acknowledgments
We thank T. Nilsen, J. A. Wise, M. Harris, E. Christian, and D. McPheeters for helpful comments on the manuscript. This work was supported by Grant MCB-9304465 from the National Science Foundation (to J.M.G.). L.V-R. was supported in part by National Institutes of Health Training Grant 5T32-GM08056.
ABBREVIATION
- ssDNA
single-stranded DNA
References
- 1.Simpson L, Emeson R B. Annu Rev Neurosci. 1996;19:27–52. doi: 10.1146/annurev.ne.19.030196.000331. [DOI] [PubMed] [Google Scholar]
- 2.Gott J M, Visomirski L M, Hunter J L. J Biol Chem. 1993;268:25483–25486. [PubMed] [Google Scholar]
- 3.Rundquist B A, Gott J M. Mol Gen Genet. 1995;247:306–311. doi: 10.1007/BF00293198. [DOI] [PubMed] [Google Scholar]
- 4.Mahendran R, Spottswood M R, Miller D L. Nature (London) 1991;349:434–438. doi: 10.1038/349434a0. [DOI] [PubMed] [Google Scholar]
- 5.Miller D, Mahendran R, Spottswood M, Ling M, Wang S, Yang N, Costandy H. In: RNA Editing: The Alteration of Protein Coding Sequences of RNA. Benne R, editor. New York: Ellis Horwood; 1993. pp. 87–103. [Google Scholar]
- 6.Mahendran R, Spottswood M S, Ghate A, Ling M-l, Jeng K, Miller D L. EMBO J. 1994;13:232–240. doi: 10.1002/j.1460-2075.1994.tb06253.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Visomirski-Robic L M, Gott J M. RNA. 1995;1:681–691. [PMC free article] [PubMed] [Google Scholar]
- 8.Thomas S M, Lamb R A, Paterson R G. Cell. 1988;54:891–902. doi: 10.1016/S0092-8674(88)91285-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cattaneo R, Kaelin K, Baczko K, Billeter M A. Cell. 1989;56:759–764. doi: 10.1016/0092-8674(89)90679-x. [DOI] [PubMed] [Google Scholar]
- 10.Paterson R G, Lamb R A. J Virol. 1990;64:4137–4145. doi: 10.1128/jvi.64.9.4137-4145.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vidal S, Curran J, Kolakofsky D. J Virol. 1990;64:239–246. doi: 10.1128/jvi.64.1.239-246.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Volchkov V E, Becker S, Volchkova V A, Ternovoj V A, Kotov A N, Netesov S V, Klenk H D. Virology. 1996;214:421–430. doi: 10.1006/viro.1995.0052. [DOI] [PubMed] [Google Scholar]
- 13.Sanchez A, Trappier S G, Mahy B W J, Peters C J, Nichol S T. Proc Natl Acad Sci USA. 1996;93:3602–3607. doi: 10.1073/pnas.93.8.3602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Benne R, Burg J V D, Brakenhoff J P J, Sloof P, Boom J H V, Tromp M C. Cell. 1986;46:819–826. doi: 10.1016/0092-8674(86)90063-2. [DOI] [PubMed] [Google Scholar]
- 15.Kable M L, Seiwert S D, Heidmann S, Stuart K. Science. 1996;273:1189–1195. doi: 10.1126/science.273.5279.1189. [DOI] [PubMed] [Google Scholar]
- 16.Cruz-Reyes J, Sollner-Webb B. Proc Natl Acad Sci USA. 1996;93:8901–8906. doi: 10.1073/pnas.93.17.8901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Blum B, Bakalara N, Simpson L. Cell. 1990;60:189–198. doi: 10.1016/0092-8674(90)90735-w. [DOI] [PubMed] [Google Scholar]
- 18.Vidal S, Curran J, Kolakofsky D. EMBO J. 1990;9:2017–2022. doi: 10.1002/j.1460-2075.1990.tb08330.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hausmann S, Jacques J-P, Kolakofsky D. RNA. 1996;2:1033–1045. [PMC free article] [PubMed] [Google Scholar]
- 20.Maslov D A, Simpson L. Cell. 1992;70:459–467. doi: 10.1016/0092-8674(92)90170-h. [DOI] [PubMed] [Google Scholar]
- 21.Sugisaki H, Takanami M. J Biol Chem. 1993;268:887–891. [PubMed] [Google Scholar]
- 22.Corell R A, Feagin J E, Riley G R, Strickland T, Guderian J A, Myler P J, Stuart K. Nucleic Acids Res. 1993;21:4313–4320. doi: 10.1093/nar/21.18.4313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miller D, Mahendran R, Spottswood M, Costandy H, Wang S, Ling M-l, Yang N. Semin Cell Biol. 1993;4:261–266. doi: 10.1006/scel.1993.1031. [DOI] [PubMed] [Google Scholar]
- 24.Chamberlain M J. Harvey Lect. 1995;88:1–21. [PubMed] [Google Scholar]