Abstract
Phosphoinositide 3-kinases (PI3Ks) are a family of lipid kinases that have been implicated in signal transduction through tyrosine kinase- and heterotrimeric G-protein-linked receptors. We report herein the cloning and characterization of p110δ, a novel class I PI3K. Like p110α and p110β, other class I PI3Ks, p110δ displays a broad phosphoinositide lipid substrate specificity and interacts with SH2/SH3 domain-containing p85 adaptor proteins and with GTP-bound Ras. In contrast to the widely distributed p110α and β, p110δ is exclusively found in leukocytes. In these cells, p110α and δ both associate with the p85α and β adaptor subunits and are similarly recruited to activated signaling complexes after treatment with the cytokines interleukin 3 and 4 and stem cell factor. Thus, these class I PI3Ks appear not to be distinguishable at the level of p85 adaptor selection or recruitment to activated receptor complexes. However, distinct biochemical and structural features of p110δ suggest divergent functional/regulatory capacities for this PI3K. Unlike p110α, p110δ does not phosphorylate p85 but instead harbors an intrinsic autophosphorylation capacity. In addition, the p110δ catalytic domain contains unique potential protein–protein interaction modules such as a Pro-rich region and a basic-region leucine-zipper (bZIP)-like domain. Possible selective functions of p110δ in white blood cells are discussed.
Phosphoinositide 3-kinases (PI3Ks) phosphorylate the 3′ OH position of the inositol ring of inositol lipids, generating phosphatidylinositol 3-phosphate, phosphatidylinositol 3,4-bisphosphate, and phosphatidylinositol 3,4,5-trisphosphate. PI3K enzymes have been identified in plants, slime molds, yeast, fruit flies, and mammals (1) and play a role in signal transduction via tyrosine kinase- and G-protein-linked receptors (2–5). In addition, PI3Ks have a function in membrane trafficking events, either constitutive or induced upon receptor stimulation (for review, see ref. 6).
Three classes of PI3Ks can be discriminated on the basis of their in vitro lipid substrate specificity. Class I PI3Ks phosphorylate phosphatidylinositol (PtdIns), phosphatidylinositol 4-phosphate, and phosphatidylinositol 4,5-bisphosphate and include p110α, β, and γ (7–10). P110α and β are closely related PI3Ks that interact with SH2/SH3-domain containing p85 adaptor proteins and with GTP–Ras (11, 12). The SH2 domains in p85 provide the heterodimeric p85/p110 PI3Ks with the capacity to interact with phosphorylated Tyr residues in receptors and other cellular proteins. P110γ, the activity of which is stimulated by G protein subunits, does not interact with p85 but instead associates with a p101 adaptor protein (13). Class II PI3Ks phosphorylate PtdIns and phosphatidylinositol 4-phosphate but not phosphatidylinositol 4,5-bisphosphate (14–16). These PI3Ks all contain a C2 domain at their C terminus. Class III PI3Ks have a substrate specificity restricted to PtdIns and are homologous to yeast Vps34p, which is involved in trafficking of proteins from the Golgi to the yeast vacuole, the equivalent of the mammalian lysosome (for review, see ref. 17).
Phosphatidylinositol 3-phosphate is constitutively present in cells and its levels are largely unaltered upon extracellular stimulation. In contrast, phosphatidylinositol 3,4-bisphosphate and phosphatidylinositol 3,4,5-trisphosphate are almost absent in quiescent cells but are produced rapidly upon stimulation by a variety of factors, suggesting a likely function as second messengers. Among the potential targets for these lipids are protein kinases such as Akt/RAC/PKB and certain protein kinase C isoforms (18–22). Akt/RAC/PKB might be upstream of targets such as p70S6k and glycogen synthase kinase 3 (23, 24). PI3Ks also affect the activity of small GTP-binding proteins such as Rac and Rab5, possibly by regulating nucleotide exchange (25, 26). Ultimately, the combination of these actions can result in cytoskeletal rearrangements, DNA synthesis/mitogenesis, cell survival, and differentiation (for review, see ref. 5).
We report the cloning of p110δ, a novel class I PI3K, and present a detailed comparison of this kinase with the other class I PI3Ks, p110α and β. Although p110δ, α, and β display many common properties, p110δ has distinct biochemical and structural features and a tissue distribution restricted to white blood cells, suggesting a distinct role and/or regulation of this new PI3K. Potential biological roles for a leukocyte-specific PI3K are discussed.
MATERIALS AND METHODS
cDNA Cloning of P110δ.
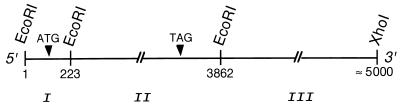
Details of the isolation of partial PI3K cDNA clones via reverse transcription-coupled PCR have been described (14, 27). A partial p110δ cDNA fragment was used to screen an oligo(dT)-primed U937 monocyte cDNA library, EcoRI–XhoI cloned in EcoRI/XhoI-digested λZAPII vector (Stratagene; ref. 27). Out of 4 × 106 clones screened, 3 primary plaques remained positive during two further rounds of screening. A schematic representation of the cDNA insert of a representative pBluescript clone (O9) is shown below.
Restriction mapping of the O9 insert revealed the absence of an internal XhoI site, and the presence of two internal EcoRI sites, 223 and 3,862 nucleotides 3′ from the EcoRI cDNA insertion site (underlined nucleotides in GAATTC is nucleotide 1). Consequently, a combined EcoRI and XhoI digest divides the O9 insert in three fragments, further named fragment I (nucleotides 1–222), II (nucleotides 223–3,861) and III (nucleotides 3,862–5,000). Both strands of fragments I and II were sequenced revealing an open reading frame spanning nucleotides 195–3,330.
Expression of P110δ in Insect Cells.
Baculovirus transfer vectors used were pVL1393 (Invitrogen) for untagged p110δ and pAcG3X (PharMingen) for GST–p110δ, which were cotransfected with BaculoGold DNA (PharMingen) in Sf9 insect cells using Lipofectin reagent (GIBCO/BRL). The p110δ coding region was cloned in these vectors in two steps. First, a BamHI–EcoRI linker (sense, 5′-GATCCCCACCATGCCCCCTGGGGTGGACTGCCCCATGG-3′; antisense, 5′-AATTCCATGGGGCAGTCCACCCCAGGGGGCATGGTGGG-3′) was inserted into BamHI–EcoRI opened vectors. This linker contains part of the sequence of EcoRI fragment I of p110δ, spanning the start codon (nucleotides 195–197; see above; underlined in the linker sequences) to the second EcoRI site (nucleotide 223; see above), and has an optimal Kozak consensus sequence surrounding the ATG (28). P110δ EcoRI fragment II was then subcloned in the EcoRI site of the linker-containing vectors, followed by selection for clones with correctly orientated inserts. Further derivatives of p110δ were made by PCR using Vent DNA polymerase (New England Biolabs). P110δ EcoRI fragment II, subcloned in the EcoRI site of pBluescript-SK (referred to as pBluescript-p110δ-EcoII) was used as a template. In these PCRs, the 3′ untranslated region of the EcoRI fragment II insert was removed. Oligonucleotides used to create R894P were as follows: sense mutagenic primer, primer 1 (mutagenic residue underlined; NdeI site in boldface type), 5′-GTGTGGCCACATATGTGCTGGGCATTGGCGATCCGCACAGCGACAACATCATGATCCG-3′, and antisense primer 2, 5′-GGCCCGGTGCTCGAGAATTCTACTGCCTGTTGTCTTTGGACACGTTGTGGGCC-3′ (stop codon underlined; XhoI site in boldface type). A parallel PCR was performed using primer 2 and a sense primer (primer 3, 5′-GTGTGGCCACATATGTGCTGGGCATTGGCG-3′; NdeI site in boldface type) leaving the wild-type p110δ kinase domain sequence intact. All PCR products were cleaved with NdeI and XhoI, subcloned into NdeI/XhoI-digested pBluescript-p110δ-EcoII, and sequenced. Correct clones were then transferred as an EcoRI cassette into EcoRI-digested pVL1393 containing the BamHI--EcoRI linker described above, followed by selection for clones with correctly orientated insert.
Lipid and Protein Kinase Assay.
Protein and lipid kinase assays and analysis of reaction products were performed as described (27, 29). Phosphoamino acid analysis was performed on a Hunter thin layer electrophoresis system (C.B.S. Scientific, Del Mar, CA) as described (30).
Antibodies and Lysis Buffers.
Monoclonal antibodies to p85α (U1 and U13) and p85β (T15) have been described (31, 32). A monoclonal Ab (I2) against p85γ was developed in our laboratory (I. Gout and M.D.W.; unpublished results). Rabbit polyclonal antiserum against glutathione S-transferase (GST)–human p85α (amino acids 5–301) was provided by P. Shepherd (University College, London). Rabbit polyclonal antibodies were raised against the following peptides: amino acids 72–88 of human p110α (used for immunoblotting), amino acids 1,054–1,068 of human p110α [provided by R. Hooshmand-Rad (Ludwig Institute for Cancer Research, Uppsala, Sweden) used for immunoprecipitation and immunoblotting], and amino acids 1,030–1,044 of p110δ (used for immunoprecipitation and immunoblotting). Both in immunoprecipitation and immunoblotting experiments, these antisera were found to be specific for the PI3Ks against which they were raised and no cross-reactivity with other known PI3Ks was observed (data not shown). Other antibodies used were 4G10 (anti-phosphotyrosine), anti-c-kit (Santa Cruz Biotechnology, sc-168), anti-SHP2 (sc-280), and anti-IRS-2 (gift of M. White, Joslin Diabetes Center, Boston, MA). Peripheral blood cells were purified over a Ficoll gradient. Neutrophil cytosol was prepared as described (33). Lysis buffer was 1% Triton X-100, 50 mM Tris⋅HCl (pH 7.4), 150 mM NaCl, 1 mM EDTA, 1 mM NaF, 1 mM NaVO3, 1 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride, aprotinin (0.27 trypsin inhibitor unit/ml), 10 μM leupeptin, 1 mM diisopropyl fluorophosphate (DIFP), and 27 μM Nα-p-tosyl-l-lysine chloromethyl ketone (TLCK). Lysis buffer used for cytokine experiments was as described (34) with the addition of 1 mM DIFP and 1 mM TLCK.
Cell Culture and Stimulations.
Murine interleukin (IL) 3-dependent cell lines Ba/F3, a pre-B cell line (35), and MC/9, a mast cell line (36), were cultured and stimulated as described (34, 37). Chemically synthesized IL-3 and IL-4 were provided by I. Clark-Lewis (University of British Columbia, Vancouver, Canada). Recombinant stem cell factor (SCF) was from R & D Systems Europe (Abingdon, Oxon, U.K.). The cytokine concentrations and duration of stimulation (2 min SCF at 50 ng/ml; 10 min IL-3 or IL-4 at 10 μg/ml) had been optimized to obtain maximal levels of tyrosine phosphorylation of receptors and cellular substrates (34, 37).
Northern Blot Analysis.
Blots of human poly(A)+ RNA (CLONTECH) were hybridized with random prime-labeled EcoRI fragment II of pBluescript-O9. Stripping and reprobing with the following probes were then subsequently performed: internal EcoRI–XhoI 2.1-kb cDNA fragment from human p110α (38) and EcoRI–XhoI 5-kb cDNA of human p110β (M.D.W., unpublished results).
RESULTS
Cloning and Primary Structure of P110δ.
Degenerate primers based on conserved amino acid sequences in PI3K catalytic core domains were used in reverse transcription-coupled PCRs (14, 27) with mRNA from the human MOLT4 T cell line. A partial cDNA, homologous to but different from other known PI3Ks, was obtained. This PCR fragment was then used as a probe to isolate full length clones from a U937 monocyte library. Sequence analysis revealed a potential open reading frame, preceded by an in-frame stop codon. The putative start codon lies in a favorable context for translation initiation (28). This open reading frame of 3,135 nucleotides encodes a protein of 1,044 amino acids with a molecular mass of 119,471 Da (Fig. 1A). Amino acid sequence comparisons revealed that this protein is most closely related to human p110β (58% overall identity), and more distantly to human p110α, p110γ, and the human Vps34p homologue (41%, 35%, and 28% overall identity, respectively). The new PI3K described herein will subsequently be referred to as p110δ.
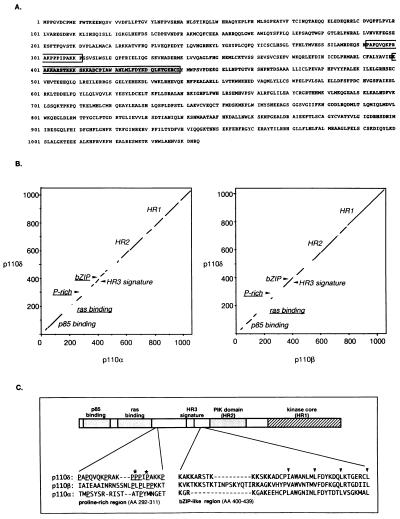
Figure 1.
(A) Translated amino acid sequence of human p110δ cDNA. The Pro-rich region and the bZIP-like domain are indicated by open and shaded box, respectively. (B) Dot plot comparison of the full-length amino acid sequence of p110δ with that of p110α and β. Nonconserved sequence motifs are underlined. Dot plot comparisons were performed with the compare program (GCG package; ref. 39). (C) Comparison of the p110δ amino acid sequence flanking HR3 with the respective homologous regions of p110α and β. Amino acid numbering is that of p110δ. In the Pro-rich region, critical Pro enabling the formation of a left-handed polyPro type II helix in p110δ are indicated with an asterisk. In the bZIP-region, conserved Leu/Val/Ile residues of the leucine-zipper region are indicated with arrowheads.
High-stringency dot plot comparison of p110δ with p110α and β showed a similar pattern of sequence conservation of all three PI3Ks (Fig. 1B). The highest homology was found in the N-terminal p85 binding region and the C-terminal homology regions (HRs) 1 and 2 [also indicated as catalytic core (HR1) and PIK domain (HR2; ref. 1)]. An additional region of high sequence homology, spanning amino acids 370–470 of p110δ, was found in between the p85 binding site and HR2. This region contains the so-called HR3 signature (WxxxLxxxIxIxDLPR/KxAxL) that is conserved in all p85-binding PI3Ks and in p110γ. It is also interesting to note from Fig. 1B that the p110 sequences seem to have diverged considerably in the region defined in p110α and β as being sufficient for Ras binding (amino acids 133–314; ref. 12).
Two additional structural motifs were identified in p110δ. The first is a Pro-rich region (Fig. 1 B and C) for which molecular modeling indicates that it can form a left-handed polyPro type II helix with the potential to interact with SH3 domains (data not shown). p110α and β lack crucial Pro residues in this region to allow a similar fold. The second motif is a basic-region, leucine-zipper (bZIP)-like domain, immediately C-terminal of HR3 (Fig. 1B and C). A bZIP region is present in both p110δ and β [and Drosophila p110 (40)], whereas the basic component of this domain is less prominent in p110α (Fig. 1C). Modeling of the p110δ ZIP region shows that its arrangement of Leu/Val/Ile residues easily accomodates the formation of a helix structure that can form a coiled-coil dimeric protein zipper complex (data not shown).
p110δ Binds the p85 Adaptor and Ras Proteins.
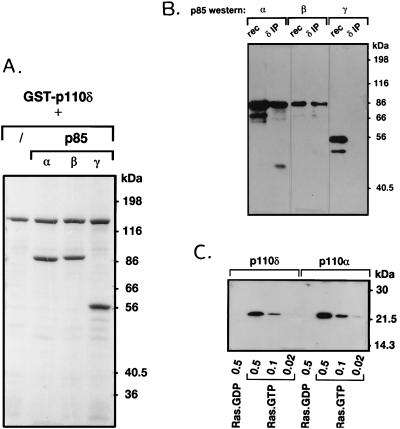
To verify the prediction from amino acid sequence comparison that p110δ might bind p85 subunits, p110δ was expressed in insect cells as a GST fusion protein, with recombinant baculoviruses encoding p85α, β, or γ [the latter is a 55-kDa bovine p85 isoform homologous to p55PIK (41)]. As is clear from Fig. 2A, all p85 adaptor subtypes efficiently copurified with GST–p110δ from coinfected cells.
Figure 2.
Interaction of p110δ with p85 and Ras. (A) Insect cells were infected with baculovirus encoding GST–p110δ alone or in combination with viruses encoding either p85α, β, or γ. After 2 days, GST–p110δ was affinity-purified from the cell lysates with glutathione-Sepharose, washed, and analyzed by SDS/PAGE and Coomassie staining. (B) p110δ was immunoprecipitated from neutrophil cytosol and probed for the presence of different p85 isoforms by Western blotting. rec, Recombinant p85 purified from Sf9 cells. (C) GST–p110α/85α and GST–p110δ/85α (0.25 μg) were incubated with the indicated amount (in μg) of GTP- or GDP-loaded V12-Ras, washed, and probed for the presence of Ras by Western blotting as described (11, 12).
The question of whether different class I p110 catalytic subunits show binding preference for different p85 adaptor proteins in vivo has not been previously addressed. With antibodies specific for p110δ, we found that both p85α and β were present in p110δ immunoprecipitates from different white blood cells (Fig. 2B shows the data for human neutrophils; note that p85γ is not expressed in leukocytes). Similar results were obtained for p110α (data not shown). In these immune complexes, a 45-kDa protein reactive with p85α antibodies was also observed (Fig. 2B). The nature of this protein is currently unclear, but it might be similar to a 45-kDa protein previously observed in p85 and p110 immunoprecipitates from various tissues (41). Also p85α and p85β immunoprecipitates blotted with antiserum against p110α or δ did not indicate a differential binding (data not shown).
p110α and β have been shown to interact with Ras–GTP (11, 12). The region required for this interaction lies between amino acids 133 and 314 of these PI3Ks (12). Despite the relatively low sequence conservation with p110α and β in this region (Fig. 1B), certain apparently critical amino acids are conserved as p110δ does interact with Ras in vitro, in a GTP-dependent manner (Fig. 2C).
p110δ Lipid Kinase Activity.
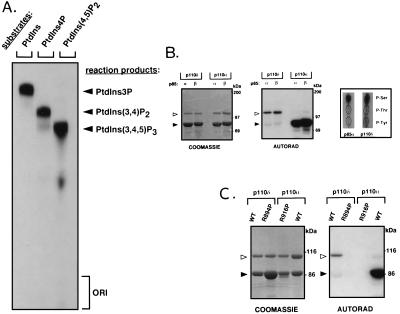
PI3K class I has been defined on the basis of the broad in vitro lipid substrate specificity of its members that can convert PtdIns, phosphatidylinositol 4-phosphate and phosphatidylinositol 4,5-bisphosphate to the respective 3′ phosphorylated derivatives (1). As is clear from Fig. 3A, p110δ was also able to phosphorylate these lipids in vitro. HPLC analysis of in vitro phosphorylated PtdIns showed that p110δ phosphorylates only at the D3 position of the inositol ring (data not shown). Thus, these data establish p110δ as a genuine class I PI3K.
Figure 3.
Enzymatic activity of p110δ. (A) In vitro lipid substrate specificity of p110δ. GST–p110δ/p85α was used in a lipid kinase assay with the indicated substrates in the presence of Mg2+. (B and C) Protein kinase activity of p110δ. Untagged p110α and δ [wild-type (WT) or kinase-defective mutants (p110α-R916P and p110δ-R894P)], in complex with p85α or β on platelet-derived growth factor receptor phosphopeptide beads, were subjected to an in vitro kinase reaction and further analyzed by SDS/PAGE, Coomassie staining, and autoradiography. Open and solid arrowheads point to p110 and p85 proteins, respectively. (B Right) Phosphoamino acid analysis of p85α and p110δ.
p110δ Protein Kinase Activity: P110δ Does not Phosphorylate p85 but Does Autophosphorylate.
The p85 adaptor protein has been shown to be a substrate for a Mn2+-dependent phosphorylation by the p110α catalytic subunit (29, 42). Interestingly, although coexpressed p85α and β were found to be good substrates for p110α protein kinase activity, they were not phosphorylated by p110δ in parallel experiments (Fig. 3B). In contrast, p110δ but not p110α autophosphorylated (Fig. 3B). Phosphoamino acid analysis showed that autophosphorylation of p110δ occurred mainly on Ser (Fig. 3B). Both the phosphorylation of p85 by p110α and the autophosphorylation of p110δ were observed to be largely Mn2+-dependent, with only very weak phosphorylation in the presence of Mg2+ (data not shown). Similar to when p85 becomes phosphorylated by p110α (29), autophosphorylation of p110δ resulted in a complete down-regulation of its lipid kinase activity (data not shown).
To exclude the possiblity that the observed phosphorylation of p110δ was due to a copurified protein kinase, a kinase-defective p110δ mutant was generated by converting Arg-894 to Pro and generating p110δ-R894P. The mutated Arg residue is located in the conserved DRx3Nx12–13DFG motif of the kinase domain, likely to be part of the catalytic loop as in protein kinases (1). A similar mutation in bovine p110α (R916P) abrogates catalytic activity (29). As is clear from Fig. 3C, purified p110δ-R894P is not autophosphorylated, indicating that the observed p110δ phosphorylation activity is intrinsic to p110δ. p110δ-R894P also no longer displayed any lipid kinase activity (data not shown).
Inhibitor Sensitivity of P110δ.
Wortmannin and LY294002 are PI3K inhibitors that have been widely used to explore the importance of this family of enzymes in cellular responses. The lipid kinase activities of p110α and δ were found to exhibit a comparable sensitivity to inhibition by wortmannin and LY294002 with an IC50 of 5 nM for wortmannin and 0.5 μM for LY294002 (data not shown). The autophosphorylation of p110δ was also inhibited by wortmannin in the nanomolar range (data not shown).
Tissue Distribution of P110δ.
The expression pattern of p110δ was investigated and compared with that of p110α and β. Whereas p110β has been demonstrated to be ubiquitous (8), the expression pattern of p110α has not yet been reported.
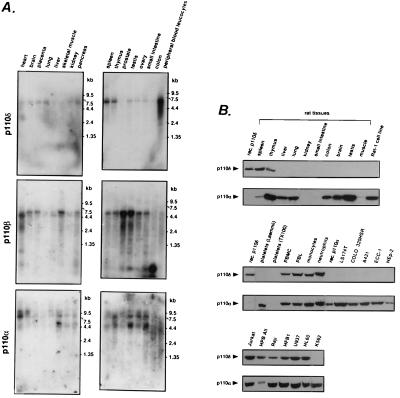
Northern blot analysis of poly(A)+ RNA from human tissues revealed an easily detectable major p110δ mRNA species of ≈6 kb in white blood cell populations, i.e., spleen, thymus, and especially in peripheral blood leukocytes (which contain all types of white blood cells, including platelets; Fig. 4A). Low levels of p110δ mRNA expression seem to be present in several other tissues although it is difficult to exclude the possibility that blood cell contamination is responsible for this signal. In contrast to p110δ, mRNAs for p110α and β were found to be more widely expressed (Fig. 4A).
Figure 4.
Tissue distribution of p110α, β, and δ. (A) Northern blot analysis of p110α, β, and δ expression. (B) Analysis of p110α and δ protein expression. Total cell lysate was loaded at 200 μg per lane. Platelets were lysed in either lysis buffer or in Laemmli gel loading buffer containing 2-mercaptoethanol. PMBC, peripheral blood mononuclear cell; PBL, peripheral blood lymphocyte.
Antibodies selective for p110α or δ were then used to examine the expression patterns of these kinases at the protein level. In rat tissues, p110δ was found in spleen and thymus but in none of the other tissues tested (Fig. 4B). p110δ was present in both primary and transformed white blood cells, independent of their differentiation stage (Fig. 4B Lower). In the primary blood cells, both the lymphoid and myeloid cell populations were positive for p110δ whereas platelets were not (Fig. 4B Middle). Both T (e.g., Jurkat and HPB All) and B (e.g., Raji and HFB1) cell lines expressed p110δ (Fig. 4B Lower). p110δ was not found in Rat-1, NIH 3T3, and Swiss 3T3 fibroblasts, LS174T and COLO 320HSR colon adenocarcinomas, A431 epidermoid carcinoma, ECC-1 endometrial carcinoma, and HEp-2 larynx carcinoma (Fig. 4B) nor in CHO Chinese hamster ovary, POC small-cell lung cancer cell line, porcine and bovine aortic endothelial cells, MDA-MB-468 breast adenocarcinoma, and primary human muscle and fibroblasts (data not shown). In conclusion, it appears that p110δ is selectively expressed in leukocytes. We would like to mention that we have found p110δ to be very sensitive to degradation, and if this is a tissue-specific phenomenon, it may explain our inability to find p110δ in some cell lines and tissues.
In contrast, p110α was found in most of the tissues and cell lines investigated, including white blood cells (Fig. 4B).
Recruitment of p110δ to Cytokine Receptors.
In leukocytes, p85-binding PI3Ks have been implicated in signaling via cytokine and complement receptors, integrins, Fc receptors, B and T cell antigen receptors, and costimulatory molecules, e.g., CD28 (for review, see refs. 2 and 3). A crucial question is whether selective coupling of p110δ to the above-mentioned signaling complexes occurs in cells that also contain other class I PI3K. We addressed this in the context of cytokine signal transduction, operative in diverse types of leukocytes.
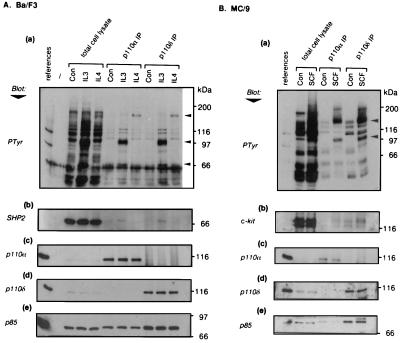
We examined the ability of IL-3, IL-4, and SCF to couple to p110δ and α in cytokine-dependent leukocyte cell lines. An identical pattern of phosphotyrosine-containing proteins, specific to the cytokine used for stimulation, was found to coprecipitate with p110α and δ antibodies (Fig. 5 Aa and Ba). In the IL-3- and IL-4-responsive Ba/F3 pre-B cell line (Fig. 5A), IL-3 treatment induced the appearance in p110α/δ immunoprecipitates of an unknown protein of 100 kDa and the 70-kDa protein tyrosine phosphatase SHP2 (Fig. 5Ab). The 170-kDa protein coprecipitated upon IL-4 stimulation (Fig. 5Aa) was shown by immunoblotting to be IRS-2, the major substrate of IL-4-induced phosphorylation in these cells (data not shown). Identical results were observed in the myeloid progenitor cells line FD-6 (data not shown). Fig. 5B shows the results of similar analyses in MC/9 mast cells. After SCF stimulation, both p110α and δ immunoprecipitates contained an unidentified 100-kDa tyrosine-phosphorylated protein and a 150-kDa protein identified as c-kit, the SCF receptor (Fig. 5 Ba and Bb). Also anti-kit receptor immunoprecipitations failed to show selective association of either p110α or δ upon ligand stimulation (data not shown). Thus, these data indicate that p110α and δ show no apparent differences in their recruitment to a variety of activated cytokine receptor complexes. In addition, the implication in cytokine signaling of at least two members of the p85-binding PI3K class reveals a previously unrecognized complication of signal transduction pathways downstream of these cytokine receptors.
Figure 5.
Involvement of p110α and δ in cytokine signaling. Ba/F3 (A) and MC/9 (B) cell lines were stimulated with the indicated cytokines. Samples from control untreated cells are labeled Con. Total cell lysates and p110α and δ immunoprecipitates were separated by SDS/PAGE to prepare duplicate blots, the references for which were p110δ/85α (a, b, and d) or p110α/85α (c and e). Immunoblotting of naive blots was performed with 4G10 (anti-phosphotyrosine) (a) and anti-p110α (c). Blots were subsequently stripped and reprobed with anti-SHP2 (Ab), anti-kit (Bb), anti-p110δ (d), and anti-p85 antibodies (e). The arrowheads indicate the positions of p170 (IRS-2), p100, and p70 (SHP2) (Aa) and of p150 (c-kit) and p100 (Bb).
DISCUSSION
Herein we describe the isolation and characterization of p110δ, a novel member of the ever expanding family of PI3Ks. Comparative analyses of the structural and biochemical characteristics of p110δ with those of p110α and β have placed p110δ in the class I PI3K. Like p110α, p110δ associates with different p85 isoforms (including p85α, β, and γ), binds Ras-GTP, exhibits a broad phosphoinositide lipid substrate specificity, and has a similar sensitivity to the inhibitors wortmannin and LY294002. An intriguing difference between the p85-binding PI3Ks is the tissue distribution of p110δ, which appears to be restricted to white blood cells, in contrast to the more ubiquitous expression of p110α and β. It is noteworthy that the same white blood cells that express p110δ also express the p110α protein (Fig. 4) and most likely p110β [based on the presence of its mRNA (Fig. 4A and ref. 8) and the presence of a p110β size protein (i.e., slightly larger than that of p110α and δ) in p85 immunoprecipitates from various white blood cells (B.V., unpublished results)]. Therefore, leukocytes are unique in that they seem to be the only cells that contain all three known members of the p85-binding PI3K subclass. When we investigated the coupling of p110δ and α to receptor complexes in lymphoid and myeloid cell lines, both PI3Ks were recruited to similar signaling complexes, the composition of which was dependent on the cytokine used. Therefore, at the level of coupling to cytokine receptor complexes, there appears to be little to distinguish the different class I PI3Ks.
These findings raise the important question as to whether these PI3Ks are functionally redundant or whether they fulfil discrete functional roles. It should be stressed that recruitment of different p110s to the same receptor complexes does not preclude the possibility of functional diversity among these PI3Ks. It is conceivable that different p85 isoforms can have nonredundant functions and may be regulated differently (32, 43). However, these differences are likely to apply to complexes of both p110α and δ because, although we have not investigated all known p85 and p110 subunits, we observed no in vivo selectivity of p110α and δ for binding either p85α or β. Moreover, potential nonredundant functions of the different p110 catalytic subunits are likely to be independent of differences between the p85 isoforms they bind. It is clear that approaches such as PI3K gene knock-out experiments will be required to resolve these issues.
Despite the similarities between p110δ and α, there are also differences and we believe these may contribute to a functional distinction between the catalytic subunits of the class I PI3Ks. The distinct protein kinase activities of p110α and δ raise the possibility of differential patterns of protein phosphorylation occurring in vivo. For example, receptor-bound p110s could induce differential phosphorylation of other proteins present in these complexes, and indeed, p85-binding PI3Ks have been shown to be capable of phosphorylating substrates other than p110 and p85 (44, 45). In addition, the p110 catalytic subunits in these complexes may themselves be subject to different regulatory modifications including phosphorylation. Clearly, the identification of the in vivo protein kinase substrates of PI3Ks is a major challenge for the future (46).
In addition to differences in protein kinase activity, divergence in the primary structure of p110 proteins may endow these subunits with discrete protein binding capacities in vivo. A discrete sequence motif such as the Pro-rich region in p110δ clearly provides this kinase with the potential for selective interaction with SH3 domain-containing proteins. Another potential protein-binding module in p110δ is the bZIP-like domain. This motif is most commonly found in transcription factors but has recently also been identified in certain protein kinases (47, 48). In transcription factors, the stretch of basic amino acids functions in DNA binding, whereas the leucine zipper is involved in transcription factor dimerization. If the bZIP domain in PI3Ks indeed functions as a binding domain, differences in this motif among the p110s (Fig. 1C) could, like for transcription factors, contribute to their binding partner specificity. An additional noteworthy observation is that the region defined in p110α and β as being sufficient for Ras binding has diverged considerably in different p85-binding PI3Ks (Fig. 1B). Although our data indicate that essential structural features for Ras binding in vitro are conserved, the possibility remains that subtle sequence differences in this part of the catalytic subunits allow a differential interaction of PI3K with different members of the family of small GTP-binding proteins in vivo, a hypothesis currently under investigation.
More specifically, in relation to a difference in functions of p110α and δ, it appears that p110δ may not be essential for cell survival or proliferation. This is based on the observations that the majority of cells exert these functions without possessing p110δ, and that transformed actively growing leukocyte cell lines lacking p110δ can be found (e.g., K562, Fig. 4B). What then might be a specific role for p110δ in white blood cells in general and in cytokine signaling in particular? A feature discriminating leukocytes from other cells in the body is their enormous circulatory and migratory capacity under the appropriate stimuli (49). Migratory responses of other cells in the organism (such as during differentiation or tissue repair) are slow, relative to those of leukocytes, and are regulated by other stimuli. It is striking that p110δ seems to be expressed in the leukocyte subpopulation capable of migrating through blood vessels, namely, the lymphoid and myeloid cells. Nonmigratory platelets, known to contain substantial amounts of p85-bound PI3K (50), do not express p110δ. Thus with the established role of PI3K in cytoskeletal rearrangements, it is tempting to speculate that p110δ uniquely contributes to the regulation of leukocyte transendothelial migration. A differential interaction with small GTP-binding proteins implicated in PI3K signaling and cytoskeletal rearrangements might add to such a specific function. This hypothesis is also consistent with the demonstrated involvement of p110δ in cytokine signaling, known to result in pleiotropic effects such as proliferation and differentiation but also cytoskeletal rearrangements and cell migration. Experiments to unveil such a potential unique function for p110δ are in progress.
Scheme I.
Acknowledgments
We thank B. Marte and E. Douville for help with lipid and phosphoamino acid analyis, and I. Gout, F. Wientjes, D. Wallis, A. Sunters, P. Shepherd, and K. Bhakoo for cell lines and tissues. We also thank S. Leevers and D. Weinkove for critically reading the manuscript and F. Pagès for the p85γ virus. B.V. is supported in part by the Belgian Fund for Scientific Research, Flanders. K.K. is supported by the Manpei Suzuki Diabetes Foundation Japan, R.S. holds a Medical Research Council Clinician Scientist Fellowship, S.V. is supported by Associazione Italiana per la Ricerca sul Cancro, and K.H. is a Visiting Scientist from the Meiji College of Pharmacy, Tokyo, Japan, supported by the Cancer Research Programme from the Ministry of Science and Education, Japan. Work in the M.J.W. laboratory was supported by a Medical Research Council Project Grant.
ABBREVIATIONS
- GST
glutathione S-transferase
- PtdIns
phosphatidylinositol
- PI3K
phosphoinositide 3-kinase
- bZIP
basic region leucine zipper
- IL
interleukin
- SCF
stem cell factor
Footnotes
References
- 1.Zvelebil M J, MacDougall L, Leevers S, Volinia S, Vanhaesebroeck B, Gout I, Panayotou G, Domin J, Stein R, Koga H, Salim K, Linacre J, Das P, Panaretou C, Wetzker R, Waterfield M D. Philos Trans R Soc London. 1996;351:217–233. doi: 10.1098/rstb.1996.0019. [DOI] [PubMed] [Google Scholar]
- 2.Stephens L R, Jackson T R, Hawkins P T. Biochim Biophys Acta. 1993;1179:27–75. doi: 10.1016/0167-4889(93)90072-w. [DOI] [PubMed] [Google Scholar]
- 3.Fry M. Biochim Biophys Acta. 1994;1226:237–268. doi: 10.1016/0925-4439(94)90036-1. [DOI] [PubMed] [Google Scholar]
- 4.Kapeller R, Cantley L C. BioEssays. 1994;8:565–576. doi: 10.1002/bies.950160810. [DOI] [PubMed] [Google Scholar]
- 5.Vanhaesebroeck B, Stein R, Waterfield M D. Cancer Surv. 1996;27:249–270. [PubMed] [Google Scholar]
- 6.De Camilli P, Emr S D, McPherson P S, Novick P. Science. 1996;271:1533–1539. doi: 10.1126/science.271.5255.1533. [DOI] [PubMed] [Google Scholar]
- 7.Hiles I D, Otsu M, Volinia S, Fry M J, Gout I, Dhand R, Panayotou G, Ruiz-Larrea F, Thompson A, Totty N F, Hsuan J J, Courtneidge S A, Parker P J, Waterfield M. Cell. 1992;70:419–429. doi: 10.1016/0092-8674(92)90166-a. [DOI] [PubMed] [Google Scholar]
- 8.Hu P, Mondino A, Skolnik E Y, Schlessinger J. Mol Cell Biol. 1993;13:7677–7688. doi: 10.1128/mcb.13.12.7677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stephens L, Smrcka A, Cooke F T, Jackson T R, Sternweiss P C, Hawkins P T. Cell. 1994;77:83–93. doi: 10.1016/0092-8674(94)90237-2. [DOI] [PubMed] [Google Scholar]
- 10.Stoyanov B, Volinia S, Hanck T, Rubio I, Loubtchenkov M, Malek D, Stoyanova S, Vanhaesebroeck B, Dhand R, Nurnberg B, Gierschik P, Seedorf K, Hsuan J J, Waterfield M D, Wetzker R. Science. 1995;269:690–693. doi: 10.1126/science.7624799. [DOI] [PubMed] [Google Scholar]
- 11.Rodriguez-Viciana P, Warne P H, Dhand R, Vanhaesebroeck B, Gout I, Fry M J, Waterfield M D, Downward J. Nature (London) 1994;370:527–532. doi: 10.1038/370527a0. [DOI] [PubMed] [Google Scholar]
- 12.Rodriguez-Viciana P, Warne P H, Vanhaesebroeck B, Waterfield M D, Downward J. EMBO J. 1996;15:2442–2451. [PMC free article] [PubMed] [Google Scholar]
- 13.Stephens L, Hawkins P T, Eguinoa A, Cooke F. Philos Trans R Soc London. 1996;351:211–215. doi: 10.1098/rstb.1996.0018. [DOI] [PubMed] [Google Scholar]
- 14.MacDougall L K, Domin J, Waterfield M D. Curr Biol. 1996;5:1404–1415. doi: 10.1016/s0960-9822(95)00278-8. [DOI] [PubMed] [Google Scholar]
- 15.Virbasius J V, Guilherme A, Czech M P. J Biol Chem. 1996;271:13304–13307. doi: 10.1074/jbc.271.23.13304. [DOI] [PubMed] [Google Scholar]
- 16.Molz L, Chen Y-W, Hirano M, Williams L T. J Biol Chem. 1996;271:13892–13899. doi: 10.1074/jbc.271.23.13892. [DOI] [PubMed] [Google Scholar]
- 17.Stack J H, Horazdovsky B, Emr S D. Annu Rev Cell Dev Biol. 1995;11:1–33. doi: 10.1146/annurev.cb.11.110195.000245. [DOI] [PubMed] [Google Scholar]
- 18.Toker A, Meyer M, Reddy K K, Falck J R, Aneja R, Aneja S, Parra A, Burns D J, Ballas L M, Cantley L C. J Biol Chem. 1994;269:32358–32367. [PubMed] [Google Scholar]
- 19.Palmer R H, Dekker L V, Woscholski R, Le Good A, Gigg R, Parker P J. J Biol Chem. 1995;270:22412–22416. doi: 10.1074/jbc.270.38.22412. [DOI] [PubMed] [Google Scholar]
- 20.Burgering B M T, Coffer P J. Nature (London) 1995;376:599–602. doi: 10.1038/376599a0. [DOI] [PubMed] [Google Scholar]
- 21.Franke T F, Yang S I, Chan T O, Datta K, Kazlauskas A, Morrison D K, Kaplan D R, Tsichlis P N. Cell. 1995;81:727–736. doi: 10.1016/0092-8674(95)90534-0. [DOI] [PubMed] [Google Scholar]
- 22.Klippel A, Reinhard C, Kavanaugh W M, Apell G, Escobedo M-A, Williams L T. Mol Cell Biol. 1996;16:4117–4127. doi: 10.1128/mcb.16.8.4117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chung J K, Grammer T C, Lemon K P, Kazlauskas A, Blenis J. Nature (London) 1994;370:71–75. doi: 10.1038/370071a0. [DOI] [PubMed] [Google Scholar]
- 24.Cross D A E, Alessi D R, Cohen P, Andjelkovich M, Hemmings B A. Nature (London) 1995;378:785–789. doi: 10.1038/378785a0. [DOI] [PubMed] [Google Scholar]
- 25.Hawkins P T, Eguinoa A, Giu R-G, Stokoe D, Cooke F T, Walters R, Wennström S, Claesson-Welsh L, Evans T, Symons M, Stephens L. Curr Biol. 1995;5:393–403. doi: 10.1016/s0960-9822(95)00080-7. [DOI] [PubMed] [Google Scholar]
- 26.Li G, D’Souza-Schorey C, Barbieri M A, Roberts R L, Klippel A, Williams L T, Stahl P D. Proc Natl Acad Sci USA. 1995;92:10207–12211. doi: 10.1073/pnas.92.22.10207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Volinia S, Dhand R, Vanhaesebroeck B, MacDougall L K, Stein R, Zvelebil M J, Domin J, Panaretou C, Waterfield M D. EMBO J. 1995;14:3339–3348. doi: 10.1002/j.1460-2075.1995.tb07340.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kozak M. J Biol Chem. 1991;266:19867–19870. [PubMed] [Google Scholar]
- 29.Dhand R, Hiles I, Panayotou G, Roche S, Fry M J, Totty N F, Truong O, Vicendo P, Yonezawa K, Kasuga M M, Courtneidge S, Waterfield M D. EMBO J. 1994;13:522–533. doi: 10.1002/j.1460-2075.1994.tb06290.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jelinek T, Weber M J. BioTechniques. 1993;15:628–630. [PubMed] [Google Scholar]
- 31.End P, Gout I, Fry M J, Panayotou G, Dhand R, Yonezawa K, Kasuga M, Waterfield M D. J Biol Chem. 1993;268:10066–10075. [PubMed] [Google Scholar]
- 32.Reif K, Gout I, Waterfield M D, Cantrell D A. J Biol Chem. 1993;268:10780–10788. [PubMed] [Google Scholar]
- 33.Wientjes F B, Hsuan J J, Totty N F, Segal A W. Biochem J. 1993;296:557–561. doi: 10.1042/bj2960557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Welham M J, Schrader J W. J Immunol. 1992;149:2772–2783. [PubMed] [Google Scholar]
- 35.Palacios R, Steinmetz M. Cell. 1985;41:727–734. doi: 10.1016/s0092-8674(85)80053-2. [DOI] [PubMed] [Google Scholar]
- 36.Nabel G, Galli S J, Dvorak A M, Dvorak H F, Cantor H. Nature (London) 1981;291:332–334. doi: 10.1038/291332a0. [DOI] [PubMed] [Google Scholar]
- 37.Welham M J, Duronio V, Schrader J W. J Biol Chem. 1994;269:5865–5873. [PubMed] [Google Scholar]
- 38.Volinia S, Hiles I, Ormondroyd E, Nizetic D, Antonacci R, Rocchi M, Waterfield M. Genomics. 1994;24:472–477. doi: 10.1006/geno.1994.1655. [DOI] [PubMed] [Google Scholar]
- 39.Devereux J, Haeberli P, Smithies O. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Leevers S J, Weinkove D, MacDougall L K, Hafen E, Waterfield M D. EMBO J. 1996;15:6584–6594. [PMC free article] [PubMed] [Google Scholar]
- 41.Pons S, Asano T, Glasheen E, Miralpeix M, Zhang Y, Fisher T C, Meyers M G, Jr, Sun X J, White M W. Mol Cell Biol. 1995;15:4453–4465. doi: 10.1128/mcb.15.8.4453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Carpenter C C, Auger K R, Duckworth B C, Hou W-M, Schaffhausen B, Cantley L C. Mol Cell Biol. 1993;13:1657–1665. doi: 10.1128/mcb.13.3.1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hartley D, Meisner H, Corvera S. J Biol Chem. 1995;270:18260–18263. doi: 10.1074/jbc.270.31.18260. [DOI] [PubMed] [Google Scholar]
- 44.Lam K, Carpenter C L, Ruderman N B, Friel J C, Kelly K L. J Biol Chem. 1994;269:24648–20652. [PubMed] [Google Scholar]
- 45.Tanti J-F, Grémaux T, Van Obberghen E, Le Marchand-Brustel Y. Biochem J. 1994;304:17–21. doi: 10.1042/bj3040017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hunter T. Cell. 1995;83:1–4. doi: 10.1016/0092-8674(95)90225-2. [DOI] [PubMed] [Google Scholar]
- 47.Ing Y L, Leung I W, Heng H H, Tsui L-C, Lassam N J. Oncogene. 1994;9:1745–1750. [PubMed] [Google Scholar]
- 48.Hirai S, Izawa M, Osada S, Spyrou G, Ohno S. Oncogene. 1996;12:641–650. [PubMed] [Google Scholar]
- 49.Springer T A. Cell. 1994;76:301–314. doi: 10.1016/0092-8674(94)90337-9. [DOI] [PubMed] [Google Scholar]
- 50.Zhang J, Zhang J, Shattil S J, Cunningham M C, Rittenhouse S E. J Biol Chem. 1996;271:6265–6272. doi: 10.1074/jbc.271.11.6265. [DOI] [PubMed] [Google Scholar]