Abstract
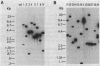
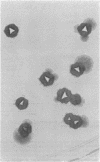
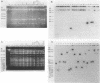
Tn5096 was constructed by inserting an apramycin resistance gene, aac(3)IV, into IS493 from Streptomyces lividans. By using conventional and pulsed-field gel electrophoresis, Tn5096 and related transposons were shown to insert into many different locations in the Streptomyces griseofuscus chromosome and in two linear plasmids. On insertion into the target site CANTg, 3 bp appeared to be duplicated. Independent transpositions were obtained by delivery of the transposon from a temperature-sensitive plasmid. The frequency of auxotrophy among cultures containing transpositions was about 0.2%.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bibb M. J., Findlay P. R., Johnson M. W. The relationship between base composition and codon usage in bacterial genes and its use for the simple and reliable identification of protein-coding sequences. Gene. 1984 Oct;30(1-3):157–166. doi: 10.1016/0378-1119(84)90116-1. [DOI] [PubMed] [Google Scholar]
- Birch A., Häusler A., Hütter R. Genome rearrangement and genetic instability in Streptomyces spp. J Bacteriol. 1990 Aug;172(8):4138–4142. doi: 10.1128/jb.172.8.4138-4142.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruton C. J., Chater K. F. Nucleotide sequence of IS110, an insertion sequence of Streptomyces coelicolor A3(2). Nucleic Acids Res. 1987 Sep 11;15(17):7053–7065. doi: 10.1093/nar/15.17.7053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chater K. F., Bruton C. J., Foster S. G., Tobek I. Physical and genetic analysis of IS110, a transposable element of Streptomyces coelicolor A3(2). Mol Gen Genet. 1985;200(2):235–239. doi: 10.1007/BF00425429. [DOI] [PubMed] [Google Scholar]
- Chung S. T. Tn4556, a 6.8-kilobase-pair transposable element of Streptomyces fradiae. J Bacteriol. 1987 Oct;169(10):4436–4441. doi: 10.1128/jb.169.10.4436-4441.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox K. L., Baltz R. H. Restriction of bacteriophage plaque formation in Streptomyces spp. J Bacteriol. 1984 Aug;159(2):499–504. doi: 10.1128/jb.159.2.499-504.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Sal G., Manfioletti G., Schneider C. The CTAB-DNA precipitation method: a common mini-scale preparation of template DNA from phagemids, phages or plasmids suitable for sequencing. Biotechniques. 1989 May;7(5):514–520. [PubMed] [Google Scholar]
- Hightower R. C., Metge D. W., Santi D. V. Plasmid migration using orthogonal-field-alternation gel electrophoresis. Nucleic Acids Res. 1987 Oct 26;15(20):8387–8398. doi: 10.1093/nar/15.20.8387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Häusler A., Birch A., Krek W., Piret J., Hütter R. Heterogeneous genomic amplification in Streptomyces glaucescens: structure, location and junction sequence analysis. Mol Gen Genet. 1989 Jun;217(2-3):437–446. doi: 10.1007/BF02464915. [DOI] [PubMed] [Google Scholar]
- Kaster K. R., Burgett S. G., Rao R. N., Ingolia T. D. Analysis of a bacterial hygromycin B resistance gene by transcriptional and translational fusions and by DNA sequencing. Nucleic Acids Res. 1983 Oct 11;11(19):6895–6911. doi: 10.1093/nar/11.19.6895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleckner N., Roth J., Botstein D. Genetic engineering in vivo using translocatable drug-resistance elements. New methods in bacterial genetics. J Mol Biol. 1977 Oct 15;116(1):125–159. doi: 10.1016/0022-2836(77)90123-1. [DOI] [PubMed] [Google Scholar]
- Larson J. L., Hershberger C. L. The minimal replicon of a streptomycete plasmid produces an ultrahigh level of plasmid DNA. Plasmid. 1986 May;15(3):199–209. doi: 10.1016/0147-619x(86)90038-7. [DOI] [PubMed] [Google Scholar]
- Leblond P., Demuyter P., Simonet J. M., Decaris B. Genetic instability and hypervariability in Streptomyces ambofaciens: towards an understanding of a mechanism of genome plasticity. Mol Microbiol. 1990 May;4(5):707–714. doi: 10.1111/j.1365-2958.1990.tb00641.x. [DOI] [PubMed] [Google Scholar]
- Lydiate D. J., Ikeda H., Hopwood D. A. A 2.6 kb DNA sequence of Streptomyces coelicolor A3(2) which functions as a transposable element. Mol Gen Genet. 1986 Apr;203(1):79–88. doi: 10.1007/BF00330387. [DOI] [PubMed] [Google Scholar]
- Olson E. R., Chung S. T. Transposon Tn4556 of Streptomyces fradiae: nucleotide sequence of the ends and the target sites. J Bacteriol. 1988 Apr;170(4):1955–1957. doi: 10.1128/jb.170.4.1955-1957.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao R. N., Richardson M. A., Kuhstoss S. Cosmid shuttle vectors for cloning and analysis of Streptomyces DNA. Methods Enzymol. 1987;153:166–198. doi: 10.1016/0076-6879(87)53053-1. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz D. C., Cantor C. R. Separation of yeast chromosome-sized DNAs by pulsed field gradient gel electrophoresis. Cell. 1984 May;37(1):67–75. doi: 10.1016/0092-8674(84)90301-5. [DOI] [PubMed] [Google Scholar]
- Siemieniak D. R., Slightom J. L., Chung S. T. Nucleotide sequence of Streptomyces fradiae transposable element Tn4556: a class-II transposon related to Tn3. Gene. 1990 Jan 31;86(1):1–9. doi: 10.1016/0378-1119(90)90107-3. [DOI] [PubMed] [Google Scholar]
- Skatrud P. L., Queener S. W. An electrophoretic molecular karyotype for an industrial strain of Cephalosporium acremonium. Gene. 1989 May 30;78(2):331–338. doi: 10.1016/0378-1119(89)90235-7. [DOI] [PubMed] [Google Scholar]
- Solenberg P. J., Burgett S. G. Method for selection of transposable DNA and characterization of a new insertion sequence, IS493, from Streptomyces lividans. J Bacteriol. 1989 Sep;171(9):4807–4813. doi: 10.1128/jb.171.9.4807-4813.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagi T. Transposition of Tn4560 in Streptomyces avermitilis. J Antibiot (Tokyo) 1990 Sep;43(9):1204–1205. doi: 10.7164/antibiotics.43.1204. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]