Abstract
The nodZ gene, which is present in various soil bacteria such as Bradyrhizobium japonicum, Azorhizobium caulinodans, and Rhizobium loti, is involved in the addition of a fucosyl residue to the reducing N-acetylglucosamine residue of lipochitin oligosaccharide (LCO) signal molecules. Using an Escherichia coli strain that produces large quantities of the NodZ protein of B. japonicum, we have purified the NodZ protein to homogeneity. The purified NodZ protein appears to be active in an in vitro transfucosylation assay in which GDP-β-fucose and LCOs or chitin oligosaccharides are used as substrates. The products of the in vitro reaction using chitin oligosaccharides as substrate were studied by using mass spectrometry, linkage analysis, and composition analysis. The data show that one fucose residue is added to C6 of the reducing-terminal N-acetylglucosamine residue. The substrate specificity of NodZ protein was analyzed in further detail, using radiolabeled GDP-β-fucose as the donor. The results show that chitin oligosaccharides are much better substrates than LCOs, suggesting that in Rhizobium NodZ fucosylates chitin oligosaccharides prior to their acylation. The free glycan core pentasaccharides of N-linked glycoproteins are also substrates for NodZ. Therefore, the NodZ enzyme seems to have an activity equivalent to that of the enzyme involved in the addition of the C6-linked fucosyl substituent in the glycan core of N-linked glycoproteins in eukaryotes. Oligosaccharides that contain only one N-acetylglucosamine at the reducing terminus are also substrates for NodZ, although in this case very high concentrations of such oligosaccharides are needed. An example is the leukocyte antigen Lewis-X, which can be converted by NodZ to a novel fucosylated derivative that could be used for binding studies with E-selectin.
Keywords: enzymology, N-acetylglucosamine, mass spectrometry, Bradyrhizobium, glycosyltransferase
The symbiotic relationship between legumes and rhizobia (i.e., Rhizobium, Bradyrhizobium, or Azorhizobium) can result in the formation of a nitrogen-fixing root organ, the nodule. The development of legume nodules is largely controlled by reciprocal signal exchange between the symbiotic partners. Legume roots secrete specific flavonoids or isoflavonoids that induce the transcription of many bacterial genes governing the early steps of this interaction (nod, nol, and noe genes). Many of these genes are involved in the synthesis and secretion of signal molecules, which are lipochitin oligosaccharides (LCOs). The chitin oligosaccharide backbone of all LCOs (also known as Nod factors) is N-acylated on the non-reducing-terminal residue. The basic structure of LCOs is synthesized by the cooperative action of NodA, NodB, and NodC (1–6). Additional gene products provide chemical decorations which, in some cases, have been shown to determine host specificity (for reviews see refs. 7–11). For instance, in a recent study (12) it was shown that transfer of the Bradyrhizobium japonicum nodZ gene to Rhizobium leguminosarum biovar viciae leads to the biosynthesis of LCOs that are fucosylated on C6 of the reducing-terminal N-acetylglucosamine, thereby extending the host range to several tropical legumes such as Macroptilium, Glycine, Vigna, and Leucaena (12). The studies of López-Lara et al. (12) and Mergaert et al. (13) suggest that nodZ encodes a fucosyltransferase.
In this study we have analyzed the function of the NodZ protein in more detail. By purifying the NodZ protein to homogeneity and subsequently analyzing its enzymatic activity in vitro we show that the protein is a glycosyltransferase that transfers fucose from GDP-β-fucose to C6 of reducing N-acetylglucosamine residues. To our knowledge, NodZ is the first example of a genetically characterized glycosyltransferase that produces oligosaccharides containing a C6 fucosyl moiety. A similar fucosyltransferase has been purified from human fibroblasts, where it is involved in the decoration of N-linked glycan moieties of proteins (14). However, the gene encoding the analogous enzyme has not yet been identified. Chitin oligosaccharides are the preferred substrates of the NodZ protein. Compounds that contain at least one N-acetylglucosamine at the reducing terminus, such as the leukocyte antigen Lewis-X (15, 16) or the free pentasaccharide core of N-linked glycoproteins, are also substrates for NodZ, although they are used less efficiently than chitin oligosaccharides. Our detailed enzymatic analysis of NodZ and the resulting fucosylated derivatives described in this paper are therefore of general importance for further studies of the function of oligosaccharides in higher organisms.
MATERIALS AND METHODS
Purification of NodZ Protein.
Cells of Escherichia coli Bl21(DE3) harboring pMP2452 or pMP2459 (Fig. 1) were grown in Luria–Bertani (LB) medium at 37°C. At OD660 = 0.3–0.4 the culture was induced with 0.1 mM isopropyl β-d-thiogalactoside (IPTG) and grown overnight. After harvesting, the cells were resuspended in 10 mM sodium phosphate buffer, pH 7.5 (P-buffer). The cells were broken by three passages through a French pressure cell [1500 psi (1 psi = 6.9 kPa)]. The NodZ protein produced forms aggregates, which were pelleted by centrifugation for 30 min at 12,000 × g. The aggregates were washed three times with P-buffer, which appeared to remove specifically the contaminating proteins. Subsequently the aggregates were dissolved in 1.85 M urea on ice. The tubes were centrifuged for 5 min at 12,000 × g to remove undissolved material. The resulting supernatant fluid contained pure NodZ, which was used directly for in vitro assays in a 10-fold dilution.
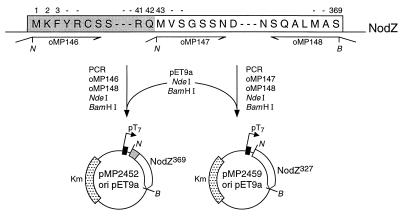
Figure 1.
Construction of plasmids. The sequence of the primers used for the PCR were as follows: oMP146, AGCTTCCATATGAAGTTCTACCGATGCAGCT; oMP148, TCAGGGGGATCCTCACGAAGCCATAAGCGCTTGCGAG; and oMP147, CGACTTCATATGGTTAGCGGTTCGAGCAATG (obtained from Isogen, Maarssen, The Netherlands).
In Vitro Activity of NodZ.
For quantification studies, time-course experiments were performed using different substrate concentrations (0.0025, 0.05, 0.075, 0.1, 0.15, 0.2, 0.4, and 0.6 mM) in the following reaction mixture: 0.86 μM GDP-β-l-[U-14C]fucose (292 mCi/mmol, Amersham; 1 mCi = 37 MBq), 0.45 mM GDP-β-l-fucose (Sigma), 0.33 mM ATP, 0.3 mM MgCl2, 0.3 mM MnCl2, 0.3 mM CaCl2, and 80 ng of NodZ protein in 10 mM sodium phosphate buffer, pH 7.5. The presence of ATP appeared to be important for activity of purified NodZ protein. When the substrate concentration was higher than 0.2 mM the concentration of GDP-l-fucose used was 0.9 mM. The total initial volume was 30 μl and for each experiment samples were taken at six time points, the interval varying from 15 sec to several hours. At each time point 4.5 μl of the mixture was diluted in 100 μl of hot water (95°C) and boiled for 10 min. The samples were treated with Dowex 1 × 8-400 ion-exchange resin (Sigma) to remove the free GDP-fucose and concentrated by vacuum evaporation to a volume of 10 μl. Stock solutions of LCOs were made by dissolving the dried compounds in water containing the detergent 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate (CHAPS; Sigma) at 1%, and these were diluted 10-fold in the reaction mixture. Control experiments using chitin pentasaccharide showed that 0.1% CHAPS does not influence the enzymatic activity of NodZ. Thin-layer chromatography (TLC) was performed using silica 60 TLC plates (Merck, Darmstadt, Germany) with 1-butanol/ethanol/water (5:3:2, vol/vol) as the mobile phase. Of each sample 2.5 μl was spotted. After overnight exposure using a phosphor screen the TLCs were visualized by using a PhosphorImager (Molecular Dynamics). For the quantification of the spots the Image Quant and Quatro Pro software were used.
For radioactive assays of the in vitro activity of NodZ, crude cell free extracts were obtained by using a French pressure cell as described previously (17). Samples equivalent to 70 μl of cell cultures were incubated in 50 μl in the presence of 25 nCi of GDP-β-l-[U-14C]fucose (Amersham), 3.3 mM ATP, and 10 μg of the chitin oligomer or other substrate at 28°C. For Lewis-X, 19 μg was used. After the reaction the samples were treated and visualized using TLC analysis as described above. Man3GlcNAc2 (M3N2), M3N2Fuc, and Lewis-X were purchased from Oxford Glycosystems (Abingdon, U.K.). Chitin oligosaccharides and chitosan oligosaccharides were purchased from Seikagaku Kogyo (Tokyo), and chitinase from Streptomyces griseus (0.01 unit per assay) was from Sigma.
HPLC Purification of Reaction Products.
Reaction products obtained from NodZ in vitro assays using the chitin pentasaccharide, trisaccharide, or M3N2 as substrate and GDP-β-l-fucose as the donor were analyzed on a Nucleosil 120–7 NH2 column (Macherey-Nagel, Düren, Germany). Before loading on the column, acetonitrile was added to the reaction mixture to a final concentration of 75% (vol/vol), and the sample was filtered through a 45-μm pore Spin X 8170 nylon membrane (Costar). Compounds were separated using an isocratic elution of 75% acetonitrile in water. Detection was by absorbance at 206 nm. Following purification the peaks were collected and concentrated by vacuum evaporation for further mass spectrometric analysis.
Fast-Atom Bombardment (FAB) MS and Collision-Induced Dissociation (CID) Tandem MS.
Positive ion mode FAB mass spectra were obtained using MS-1 of a JEOL JMS-SX/SX102A tandem mass spectrometer operating at an accelerating voltage of 10 kV. The FAB gun was operating at an accelerating voltage of 6 kV with an emission current of 10 mA and xenon was used as the bombarding gas. Spectra were scanned at a speed of 30 s for the full mass range specified by the accelerating voltage used, and they were recorded and averaged using a Hewlett–Packard HP9000 data system running JEOL complement software.
CID mass spectra were recorded using the same instrument, introducing air into the collision cell in the third field-free region at a pressure sufficient to reduce the parent ion to one third of its original intensity. Native samples were dissolved in 30 μl of distilled water, while permethylated oligosaccharides were redissolved in 10 μl of methanol. Aliquots (5 μl) of the sample solution were loaded into a matrix of thioglycerol.
Gas Chromatography–Mass Spectrometry (GC-MS).
To establish the composition and linkages, the oligosaccharides were converted to their trimethylsilyl (TMS) methyl glycosides by methanolysis and trimethylsilylation as described (18) and were separated and identified by using GC-MS, or were converted to their partially methylated alditol acetates (PMAAs) by permethylation, hydrolysis, reduction, and acetylation (18), and analyzed by using GC-MS. GC-MS analyses were performed using a Fisons MD800 mass spectrometer fitted with a Carlo Erba GC8060 gas chromatograph, an on-column injector, and using helium as the carrier gas. Monosaccharide derivatives were separated on a DB-5MS column (0.25 mm × 30 m; J & W Scientific, Folsom, CA). TMS methyl glycosides were injected directly from solution in the TMS reagent (1 μl injected) and separated using the following temperature program: 110°C for 2 min, then ramping at 30°C/min to 140°C, holding for 2 min, then ramping at 4°C/min to 180°C, holding for 30 min, then finally ramping at 30°C/min to 250°C and holding for 10 min. PMAAs were dissolved in dichloromethane before injection (1 μl injected) and separated using the following temperature program: 50°C for 2 min, then ramping at 40°C/min to 130°C, holding for 2 min, then ramping at 4°C/min to 230°C, and holding for 15 min. Mass spectra were recorded under conditions of electron ionization in the positive ion mode with an electron energy of 70 eV, and using linear scanning from m/z 50 to m/z 350 over 1 s.
RESULTS
Overproduction, Purification, and in Vitro Activity of NodZ Protein.
Suitable restriction sites for cloning the B. japonicum nodZ gene product in expression vector pET9a were introduced by PCR (Fig. 1). Two possible starting codons were considered (12), leading to construct pMP2452 and pMP2459 (Fig. 1). E. coli BL21(DE3) harboring pMP2452 (with the largest open reading frame, 369 amino acid residues) produced isopropyl β-d-thiogalactoside-inducible protein, which migrated as an approximately 41-kDa protein on sodium dodecyl sulfate/polyacrylamide gel electrophoresis (SDS/PAGE) (Fig. 2A, lane 3). This apparent molecular mass is in agreement with the expected size of 369 amino acid residues (12). Pure and active NodZ protein was obtained from NodZ protein aggregates produced by E. coli (pMP2452), as described in Materials and Methods. The results show that out of the protein bodies a single band of the expected apparent molecular mass was obtained as judged by SDS/PAGE and silver staining (Fig. 2A, lanes 4 and 5). To show in vitro transfucosylating activity of the purified NodZ protein, purified native NodZ protein from the E. coli(pMP2452) inclusion bodies was incubated in the presence of GDP-β-l-[U-14C]fucose and chitin trisaccharide as candidate fucosyl acceptor substrate. A radioactive reaction product was formed and detected on TLC (Fig. 2B, lane 1); it migrates at the same position as the labeled reaction product obtained when soluble protein fractions isolated from induced E. coli(pMP2452) were assayed for NodZ activity (Fig. 2B, lane 2). These reaction products run at positions different from the reference chitin oligosaccharides (Fig. 2B, lane 3). These results suggest that these reaction products contain an additional fucosyl group and, since only one reaction product is formed, this suggests that only one fucosyl moiety is substituted per substrate molecule. Similar results were obtained when the construct pMP2459 was used instead of pMP2452, showing that the 42 N-terminal amino acids of the longest open reading frame are not necessary for activity of the protein (data not shown).
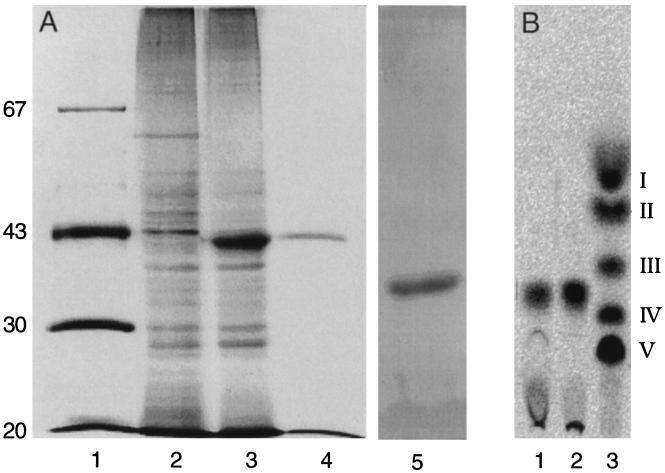
Figure 2.
Purification of active NodZ protein. (A) Samples from the purification steps were analyzed by PAGE using silver staining for detection of protein. Lane 1, marker proteins with the molecular masses indicated in kDa; lane 2, crude protein extract of E. coli strain BL21(DE3) harboring pET9A; lane 3, crude protein extract of E. coli strain BL21(DE3) harboring pMP2452; lane 4, protein fraction obtained after the last purification step (the quantity applied is equivalent to the yield from the sample in lane 3); and lane 5, test of purity of 1 μg of purified NodZ protein. (B) TLC analysis of reaction products resulting from in vitro activity of purified NodZ protein. Lane 1, reaction product of chitin trisaccharide incubated with the pure protein preparation analyzed in A, lane 4; lane 2, reaction product of chitin trisaccharide incubated with the crude protein preparation of A, lane 3; lane 3, standard of radiolabeled chitin oligosaccharides (chain-length V to II) and N-acetylglucosamine (I) as described by Kamst et al. (3).
Substrate Specificity of NodZ Protein.
GDP-α-fucose and GDP-β-mannose were tested as donors for the fucosyltransferase activity to chitin pentasaccharide. No conversion of the chitin pentasaccharide was detected when these compounds were tested as fucosyl donors and HPLC was used for detection of fucosylated derivatives (legend to Fig. 4). These results indicate that the NodZ enzyme is highly specific for GDP-β-fucose as the donor substrate. To analyze the specificity of NodZ protein for the acceptor substrates, enzymatic reactions in the presence of GDP-β-l-[U-14C]fucose and various oligosaccharides were carried out. Chitin oligosaccharides, N-acetylglucosamine, LCOs, and oligosaccharides that contain at least one N-acetylglucosamine at the reducing terminus, such as the leukocyte antigen Lewis-X or the M3N2 glycan core of N-linked glycoproteins, were tested.
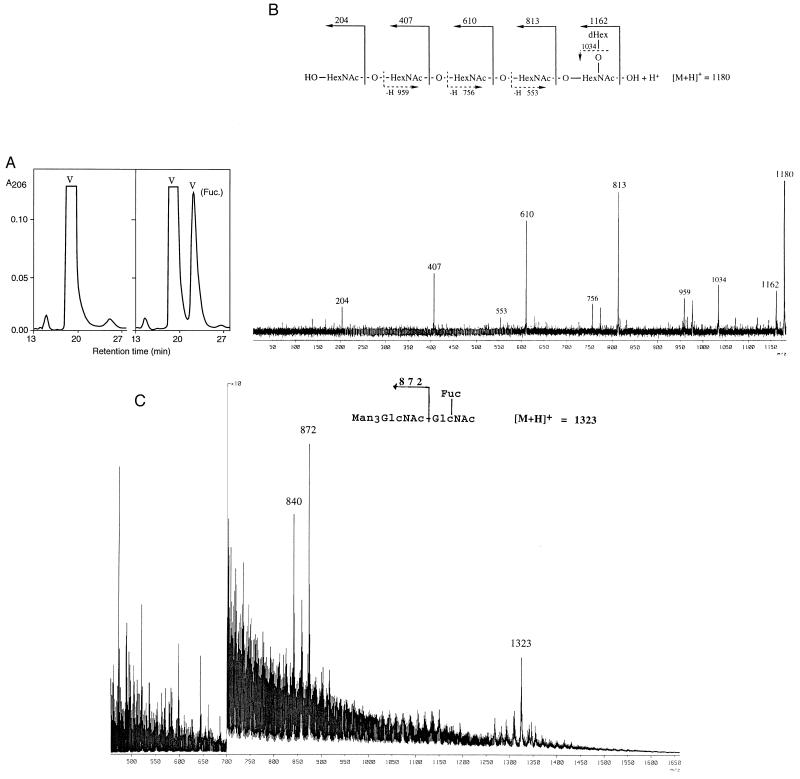
Figure 4.
(A) Conversion of chitin pentasaccharide to the fucosylated derivative as monitored by HPLC (NH2-silica column). Comparable experiments in which GDP-β-mannose and GDP-α-fucose were added instead of GDP-β-fucose did not yield a reaction product (data not shown). The conversion of M3N2, which migrates with a retention time of 24 min, resulted in a NodZ-dependent peak with a retention time of 27 min. (B) FAB CID tandem mass spectrum of the pseudomolecular ion of the product obtained after incubation with the pentasaccharide precursor GlcNAc5 with NodZ. (C) FAB mass spectrum obtained after permethylation of the product obtained on incubation with the pentasaccharide M3N2. The ion at m/z 840 results from that at m/z 872 by elimination of methanol from C3 of the charge-bearing GlcNAc residue, consistent with C3 having a methoxy group substituent.
When purified NodZ protein and GDP-β-l-[U-14C]fucose were incubated with N-acetylglucosamine (GlcNAc) or chitin fragments (di-, tri-, tetra-, penta-, and hexameric forms of N-acetylglucosamine), radioactive reaction products were detected on TLC (Fig. 3A, lanes 3–8). These reaction products run at positions only slightly different from those of the nontreated reference compounds, suggesting that these reaction products contain an additional fucosyl group. In the case of GlcNAc two reaction products were detected. The TLC analysis indicates that the major reaction product migrates with free fucose, and that the minor reaction product is a fucosylated derivative of GlcNAc. The production of fucose is probably the result of a specific hydrolysis of GDP-fucose by the NodZ protein, indicating that GlcNAc is a very poor acceptor substrate. O-Acetylchitin pentasaccharide, produced by the use of the transacetylase NodL (20), was also tested as an acceptor substrate. This derivative contains an O-acetyl on C6 of the non-reducing-terminal GlcNAc residue. The results (not shown) demonstrate that O-acetylated derivatives are also substrates for NodZ. For the assays of LCOs we made use of samples which were purified from R. leguminosarum biovar viciae. TLC analysis of LCOs which were tested in the NodZ enzymatic assay shows that they can be used as acceptor substrates (Fig. 3B, lanes 1 and 2). We were interested in determining whether compounds of animal origin, such as oligosaccharides that contain at least one N-acetylglucosamine at the reducing terminus, could be recognized as acceptors by the bacterial fucosyltransferase. As shown in Fig. 3C, lanes 1 and 3, both M3N2 and Lewis-X can be used as substrates for NodZ transfucosylating activity. The following substrates were found not to be active as fucosyl acceptor substrates: (i) chitosan oligosaccharides (i.e., the de-N-acetylated form of chitin oligosaccharides), (ii) cellulose oligosaccharides, and (iii) a derivative of M3N2 that already contains a fucosyl moiety at position C6 of the reducing GlcNAc (M3N2F). These results indicate that NodZ is specific for the C6 position of reducing-terminal GlcNAc residues.
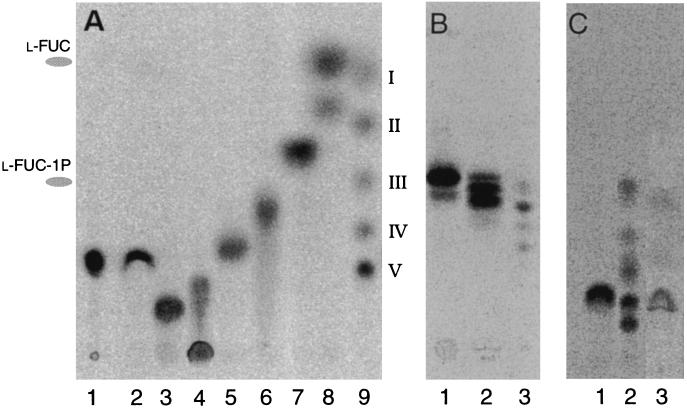
Figure 3.
TLC analysis of reaction products of NodZ protein with various substrates and GDP-β-l-[U-14C]fucose. (A) Silica TLC. Lanes: 1, standard GDP-β-l-[U-14C]fucose; 2, incubation of the negative control extract shown in Fig. 2A, lane 2, with chitin pentasaccharide; 3, chitin hexasaccharide; 4, chitin pentasaccharide; 5, chitin tetrasaccharide; 6, chitin trisaccharide; 7, chitin disaccharide; 8, N-acetylglucosamine; and 9, standard radiolabeled chitin oligosaccharides (chain-length V to II) and N-acetylglucosamine (I) as described by Kamst et al. (3). Indicated at the left is the migration of the reference compounds l-fucose and l-fucose 1-phosphate. (B) C18-silica TLC. Lanes: 1, LCO NodRlv-V (C18:4, Ac); 2, mixture of LCOs NodRlv-IV; 3, standard of nonfucosylated 14C-labeled LCOs NodRlv-V and NodRlv-IV (ref. 19; nomenclature described in ref. 12). (C) Silica TLC. Lanes: 1, Man3GlcNAc2 (M3N2); 2, standard as shown in lane 9 of A; and 3, Lewis-X.
Quantitative Analysis of NodZ Protein Substrate Specificity.
To obtain indications about the in vivo fucosyl-acceptor substrates for NodZ, a quantitative analysis of substrate specificity was performed. Initial reaction rates of transfucosylating activity were determined for various chitin oligosaccharides, LCOs, and for the core glycan moiety of N-linked glycoproteins (M3N2). Initial reaction rates for 0.1 mM and 0.4 mM substrates were determined (Table 1). The results show that (i) the reaction rates for chitin oligosaccharides are much higher than those for LCOs or M3N2, and (ii) the most efficient substrates are the penta- and hexasaccharide forms of chitin. Competition experiments in which several possible substrates were mixed in equimolar concentrations and treated with NodZ confirmed that LCOs are much less efficient substrates than chitin oligosaccharides (data not shown). Lewis-X appears to be a very poor substrate, and kinetic studies could therefore be performed only at concentrations higher than 2.4 mM, a concentration at which a reaction velocity of 1.4 × 10−12 mol/min was determined. Further kinetic studies were performed using the chitin pentasaccharide. The initial reaction rates for this substrate were determined in triplicate at various concentrations and the resulting reaction rates were analyzed using a Lineweaver–Burk plot. Using nonlinear regression, we determined a Km value of 0.12 ± 0.02 mM.
Table 1.
Reaction velocities of NodZ protein with various fucosyl-acceptor substrates at 0.1 mM or 0.4 mM substrate concentration
| Substrate | Velocity, nmol/min
|
|
|---|---|---|
| 0.1 mM | 0.4 mM | |
| Chitin hexasaccharide | 3.9 | 11.7 |
| Chitin pentasaccharide | 3.3 | 7.8 |
| Chitin tetrasaccharide | 1.2 | 4.5 |
| Man3GlcNAc2 | 0.042 | 0.1 |
| NodRlv-V (C18:4, Ac) | 0.0075 | 0.03 |
Eighty nanograms of NodZ protein was used for each assay.
Complete Chemical Analysis of in Vitro Reaction Products.
The products obtained following incubation of NodZ protein with the GDP-β-l-fucose donor and the chitin trisaccharide, pentasaccharide, or M3N2 were separated on reversed-phase high-performance liquid chromatography (HPLC). Each chromatogram had one fast and one slow eluting peak as exemplified for the chitin pentasaccharide (Fig. 4A). The fast eluting peaks have retention times identical to those of the unmodified oligosaccharide standards. The slow eluting peak for the M3N2 compound has a retention time identical to that of the commercially available M3N2F standard. The HPLC peak fractions were studied using positive ion mode FAB MS.
The results of the analyses of the fast eluting peaks show that these correspond to unmodified oligosaccharide substrates (data not shown). The slow eluting fraction obtained from the chitin pentasaccharide incubation (Fig. 4A) yielded a mass spectrum containing signals at m/z 1180 and m/z 1202, corresponding to [M + H]+ and [M + Na]+ pseudomolecular ions for a hexasaccharide with composition deoxyHex1HexNAc5. The CID mass spectrum of the ion at m/z 1180 contains signals of high intensity at m/z 813, m/z 610, and m/z 407, corresponding to B-ions, and signals of lower intensity at m/z 959, m/z 756, and m/z 553, corresponding to Z-ions (Fig. 4B), showing that a deoxyhexose residue is attached to the reducing-terminal HexNAc residue. The CID mass spectrum of the [M + H]+ pseudomolecular ion at m/z 1446 in the spectrum of the permethylated derivative of this compound has signals at m/z 1414, m/z 995, m/z 750, m/z 505, and m/z 260. These signals correspond to B5 (deoxyHex1HexNAc5), B4 (HexNAc4), B3 (HexNAc3), B2 (HexNAc2), and B1 (HexNAc1). This supports our assignment of a structure in which the deoxyhexose residue is attached to the reducing-terminal HexNAc residue.
The slow eluting fraction from the chitin trisaccharide incubation yielded a mass spectrum containing signals at m/z 774 and m/z 796, corresponding to [M + H]+ and [M + Na]+ pseudomolecular ions for a deoxyHex1HexNAc3 tetrasaccharide. The CID mass spectrum of the ion at m/z 774 contains abundant signals at m/z 407 and m/z 204, corresponding to B-ions, together with signals of lower intensity at m/z 553 and m/z 350, corresponding to Z-ions, again allowing us to assign the site of attachment of the deoxyhexose residue to be the reducing-terminal HexNAc residue. Following permethylation, the CID mass spectrum of the [M + H]+ pseudomolecular ion (m/z 956) was recorded. It contains abundant signals at m/z 924, m/z 505, and m/z 260, corresponding to a deoxyHex1HexNAc3 B3-ion, a HexNAc2 B2-ion, and a HexNAc B1-ion, respectively, confirming the presence of the deoxyhexose residue on the reducing-terminal HexNAc residue.
Since the activated monosaccharide donor used was GDP-fucose, the deoxyhexose residue was expected to be fucose. Composition analysis was carried out to confirm this. Fraction 2 from the incubations of the β-fucose donor with GlcNAc3 and GlcNAc5, together with authentic fucose and rhamnose standards, were subjected to methanolysis and trimethylsilylation, and the resulting monosaccharide derivatives were separated and analyzed using capillary GC-MS. Both samples yielded abundant peaks corresponding to TMS methyl glycosides, eluting with retention times and peak patterns corresponding to those typically obtained from standard fucose, while peaks with retention times corresponding to standard rhamnose were completely absent.
The linkage position of the fucose to the reducing-terminal residue was determined following preparation of PMAAs from GlcNAc3Fuc and GlcNAc5Fuc. Identification of the resulting monosaccharide derivatives was carried out using capillary GC-MS. Both compounds yielded three peaks, which were identified as corresponding to non-reducing-terminal HexNAc, 4-substituted HexNAc, and 4,6-disubstituted HexNAc. These data allow us to define the site of fucosylation as C6 of the reducing-terminal GlcNAc residue in both the GlcNAc3 and GlcNAc5 experiment.
The structure of the compound in the fast eluting peak of the M3N2 incubation was determined mass spectrometrically following permethylation to improve sensitivity and fragmentation characteristics. The mass spectrum obtained was identical to that of permethylated standard M3N2Fuc (data not shown) and contained an [M + H]+ pseudomolecular ion at m/z 1323, corresponding to the fully methylated species, and, importantly, at m/z 872 an intense B-type fragment ion corresponding to Man3GlcNAc1+, which demonstrates that the site of attachment of the deoxyhexose residue is the reducing-terminal GlcNAc residue (Fig. 4C).
DISCUSSION
As the result of many recent studies of LCOs we are now able to understand the general pathway leading to the biosynthesis of these signal molecules. One of the aims of the present study was to analyze the fucosylation step in the biosynthesis of LCOs in more detail. Our study demonstrates that the product of the gene nodZ is a fucosyltransferase able to produce a derivative of a chitin oligosaccharide that is fucosylated on C6 of the reducing-terminal GlcNAc moiety. A comparison of the enzymatic activity with various other possible substrates shows that, although chitin oligosaccharides are the preferred substrates for NodZ, other oligosaccharides that contain an unsubstituted GlcNAc residue at the reducing terminus can also act as substrates. One of the best alternatives to chitin oligosaccharides is the M3N2 glycan, the structure of which is common to the core of all N-linked glycans. A fucosyl group can be found on C6 of the reducing-terminal GlcNAc in N-linked glycoprotein glycans, and an enzymatic activity similar to that of NodZ has been reported to be present in eukaryotic organisms (14, 21, 22). The identification of the corresponding gene might be assisted by the results presented in this paper. It would be of interest to compare directly the substrate specificity of NodZ with that of its eukaryotic counterpart. We have also tested the activity of NodZ with the substrate Lewis-X, one of the ligands for the leukocyte-binding E-selectin (15, 16). Using NodZ, we were able to obtain a fucosylated derivative of Lewis-X which has not yet been described. This new derivative (or an analogous fucosylated derivative of sialyl Lewis-X) could be useful for further analysis of the specificity of binding to the E-selectin protein.
Comparison of the in vitro selectivity of the NodZ enzyme for various substrates indicates that in Rhizobium NodZ is active before the acylation step of chitin oligosaccharides. This is in contrast to the results obtained with the transsulfation enzyme NodH, which is active after the acylation step, and which produces an LCO modified at the C6 position of the reducing-terminal GlcNAc (23). Knowledge of the various steps in LCO biosynthesis has already proven to be vital to the understanding of the processes of signal recognition in the rhizobial–host plant interaction (e.g., see refs. 8 and 24). In addition, this knowledge can now be used to obtain various derivatives of LCOs (e.g., radiolabeled derivatives) which will be of importance in the biochemical study of putative plant LCO receptors. Recent studies indicate that LCOs or chitin oligosaccharides might also play a role in developmental processes other than root nodulation. For instance, the results of Semino et al. (25, 26) indicate that chitin oligosaccharides are also produced during the embryogenic development of vertebrates, such as zebrafish. The role of the DG42 gene product, a homologue of chitin synthases, in the production of these chitin oligosaccharides has recently been disputed in refs. 27 and 28. We are currently involved in studies of the occurrence of LCOs or chitin oligosaccharides in eukaryotes, including plants and vertebrates. For these studies, the availability of enzymes that can be used to radiolabel such molecules plays an indispensable role. Preliminary results indicate that NodZ is ultimately suited for such studies, since control experiments show that, using radiolabeled GDP-β-fucose, we are able to specifically detect chitin oligosaccharides at quantities as low as 1 pmol (data not shown).
Acknowledgments
We thank Prof. D.H. van den Eijnden (Amsterdam) for stimulating discussions. This work was supported by contracts from the European Union, BIO2-CT92-5112 (fellowship to I.M.L.-L.) and BIO2-CT93–0400 (DG12 SSMA), the Netherlands Foundation for Chemical Research (SON) (J.T.O. and G.V.B.), the Netherlands Organization for the Advancement of Pure Research (NWO-PIONIER grant awarded to H.P.S.). C.Q. was supported by a Marie Curie fellowship from the European Union for a research project at Leiden University and from the General Direction of Academic Staff Affairs, Universidad Nacional Autónoma de México.
ABBREVIATIONS
- CID
collision-induced dissociation
- FAB
fast-atom bombardment
- LCO
lipochitin oligosaccharide
- M3N2
Man3GlcNAc2
- PMAAs
partially methylated alditol acetates
- TMS
trimethylsilyl
References
- 1.Atkinson E M, Palcic M M, Hindsgaul O, Long S R. Proc Natl Acad Sci USA. 1994;91:8418–8422. doi: 10.1073/pnas.91.18.8418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Geremia R A, Mergaert P, Geelen D, Van Montagu M, Holsters M. Proc Natl Acad Sci USA. 1994;91:2669–2673. doi: 10.1073/pnas.91.7.2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kamst E, van der Drift K M G M, Thomas-Oates J E, Lugtenberg B J J, Spaink H P. J Bacteriol. 1995;177:6282–6285. doi: 10.1128/jb.177.21.6282-6285.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mergaert P, D’Haeze W, Geelen D, Promé D, Van Montagu M, Geremia R, Promé J-C, Holsters M. J Biol Chem. 1995;270:29217–29223. doi: 10.1074/jbc.270.49.29217. [DOI] [PubMed] [Google Scholar]
- 5.Röhrig H, Schmidt J, Wieneke U, Kondorosi E, Barlier I, Schell J, John M. Proc Natl Acad Sci USA. 1994;91:3122–3126. doi: 10.1073/pnas.91.8.3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Spaink H P, Wijfjes A H M, van der Drift K M G M, Haverkamp J, Thomas-Oates J E, Lugtenberg B J J. Mol Microbiol. 1994;13:821–831. doi: 10.1111/j.1365-2958.1994.tb00474.x. [DOI] [PubMed] [Google Scholar]
- 7.Carlson R W, Price N P J, Stacey G. Mol Plant–Microbe Interact. 1994;7:684–695. doi: 10.1094/mpmi-7-0684. [DOI] [PubMed] [Google Scholar]
- 8.Dénarié J, Cullimore J. Cell. 1993;74:951–954. doi: 10.1016/0092-8674(93)90717-5. [DOI] [PubMed] [Google Scholar]
- 9.Downie J A. Trends Microbiol. 1994;2:318–324. doi: 10.1016/0966-842x(94)90448-0. [DOI] [PubMed] [Google Scholar]
- 10.Schultze M, Kondorosi A. World J Microbiol Biotechnol. 1996;12:137–147. doi: 10.1007/BF00364678. [DOI] [PubMed] [Google Scholar]
- 11.Spaink H P. Annu Rev Phytopathol. 1995;33:345–368. doi: 10.1146/annurev.py.33.090195.002021. [DOI] [PubMed] [Google Scholar]
- 12.López-Lara I M, Blok-Tip L, Quinto C, Garcia M L, Stacey G, Bloemberg G V, Lamers G E M, Lugtenberg B J J, Thomas-Oates J E, Spaink H P. Mol Microbiol. 1996;21:397–408. doi: 10.1046/j.1365-2958.1996.00644.x. [DOI] [PubMed] [Google Scholar]
- 13.Mergaert P, D’Haeze W, Fernández-López M, Geelen D, Goethals K, Promé J-C, Van Montagu M, Holsters M. Mol Microbiol. 1996;21:409–419. doi: 10.1046/j.1365-2958.1996.6451366.x. [DOI] [PubMed] [Google Scholar]
- 14.Voynow J A, Kaiser R S, Scanlin T F, Glick M C. J Biol Chem. 1991;266:21572–21577. [PubMed] [Google Scholar]
- 15.Hakomori S. Histochem J. 1992;24:771–776. doi: 10.1007/BF01046348. [DOI] [PubMed] [Google Scholar]
- 16.Varki A. Proc Natl Acad Sci USA. 1994;91:7390–7397. doi: 10.1073/pnas.91.16.7390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bloemberg G V, Thomas-Oates J E, Lugtenberg B J J, Spaink H P. Mol Microbiol. 1994;11:793–804. doi: 10.1111/j.1365-2958.1994.tb00357.x. [DOI] [PubMed] [Google Scholar]
- 18.López-Lara I M, van den Berg J D J, Thomas-Oates J E, Glushka J, Lugtenberg B J J, Spaink H P. Mol Microbiol. 1995;15:627–638. doi: 10.1111/j.1365-2958.1995.tb02372.x. [DOI] [PubMed] [Google Scholar]
- 19.Spaink H P, Aarts A, Stacey G, Bloemberg G V, Lugtenberg B J J, Kennedy E P. Mol Plant–Microbe Interact. 1992;5:72–80. doi: 10.1094/mpmi-5-072. [DOI] [PubMed] [Google Scholar]
- 20.Bloemberg G V, Kamst E, Harteveld M, van der Drift K M G M, Haverkamp J, Thomas-Oates J E, Lugtenberg B J J, Spaink H P. Mol Microbiol. 1995;16:1123–1136. doi: 10.1111/j.1365-2958.1995.tb02337.x. [DOI] [PubMed] [Google Scholar]
- 21.Longmore G D, Schachter H. Carbohydr Res. 1982;100:365–392. doi: 10.1016/s0008-6215(00)81049-6. [DOI] [PubMed] [Google Scholar]
- 22.Staudacher E, Altmann F, Glossl J, Marz L, Schachter H, Kamerling J P, Hård K, Vliegenthart J F. Eur J Biochem. 1991;199:745–751. doi: 10.1111/j.1432-1033.1991.tb16179.x. [DOI] [PubMed] [Google Scholar]
- 23.Schultze M, Staehelin C, Röhrig H, John M, Schmidt J, Kondorosi E, Schell J, Kondorosi A. Proc Natl Acad Sci USA. 1995;92:2706–2709. doi: 10.1073/pnas.92.7.2706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Spaink H P. Crit Rev Plant Sci. 1996;15:559–582. [Google Scholar]
- 25.Semino C E, Robbins P W. Proc Natl Acad Sci USA. 1995;92:3498–3501. doi: 10.1073/pnas.92.8.3498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Semino C E, Specht C A, Raimondi A, Robbins P W. Proc Natl Acad Sci USA. 1996;93:4548–4553. doi: 10.1073/pnas.93.10.4548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meyer M F, Kreil G. Proc Natl Acad Sci USA. 1996;93:4543–4547. doi: 10.1073/pnas.93.10.4543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.DeAngelis P L, Acbyuthan A M. J Biol Chem. 1996;271:23657–23660. doi: 10.1074/jbc.271.39.23657. [DOI] [PubMed] [Google Scholar]