Abstract
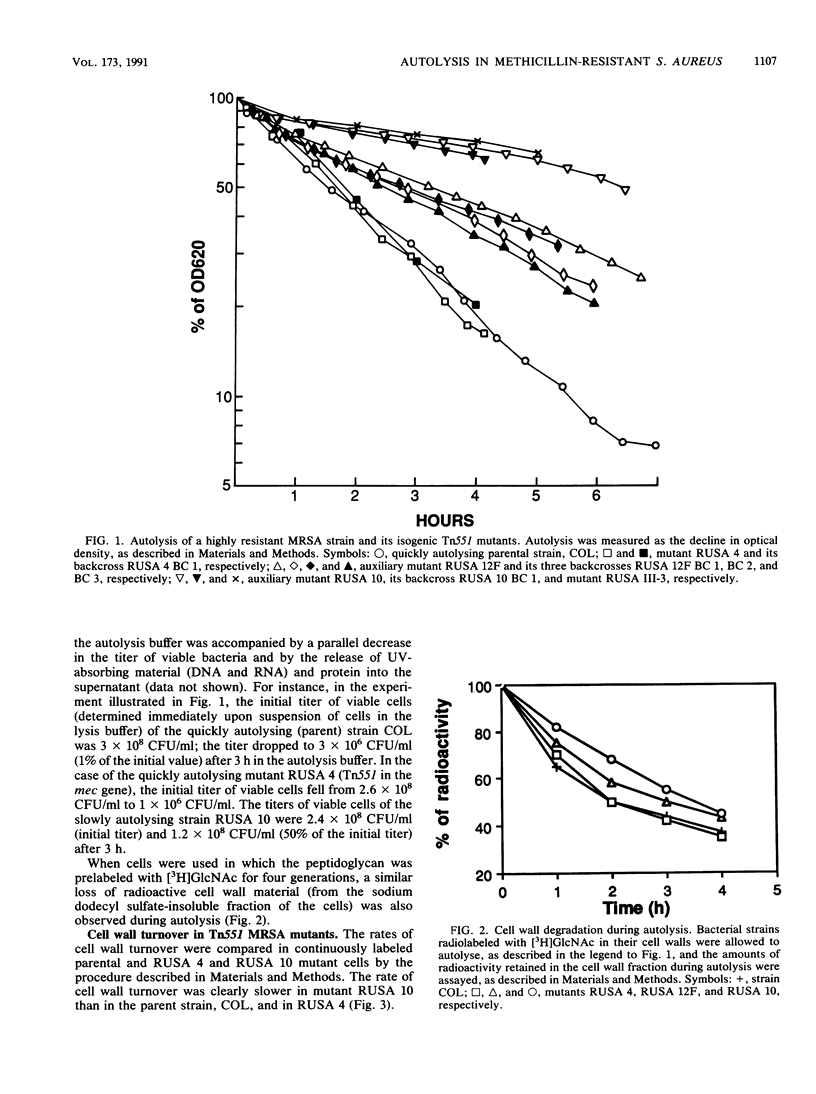
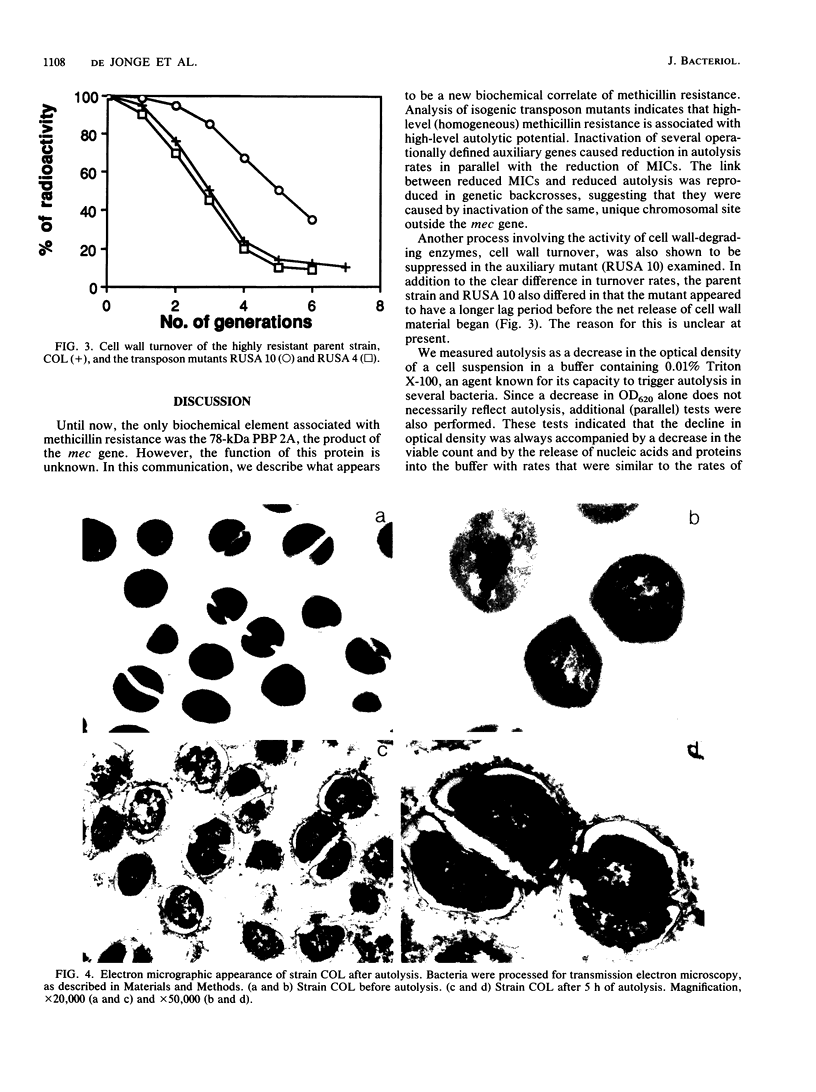
Isogenic Tn551 mutants of a highly and uniformly methicillin-resistant strain of Staphylococcus aureus were tested for their rates of autolysis and cell wall degradation in buffer and for cell wall turnover during growth. The normal (relatively fast) autolysis and turnover rates of the parent strain were retained in a Tn551 mutant in which the insert was located within the mec gene and which produced undetectable levels of penicillin-binding protein 2A. On the other hand, autolysis and cell wall turnover rates were greatly reduced in auxiliary mutants, i.e., mutants in which the transposon caused conversion of the high-level and uniform resistance of the parent strain to a variety of distinct heterogeneous expression types and greatly decreased resistance levels. All of these mutants contained an intact mec gene and produced normal amounts of penicillin-binding protein 2A, and one of the mutations was located in the femA region of the staphylococcal chromosome (B. Berger-Bachi, L. Barberis-Maino, A. Strassle, and F. H. Kayser, Mol. Gen. Genet. 219:263-269, 1989). Autolysis rates were related to the degree of residual methicillin resistance and to the sites of Tn551 insertion. Fast cell wall turnover may help expression of high-level methicillin resistance by providing a mechanism for the excision of abnormal (and potentially lethal) structural elements of the cell wall synthesized by the bacteria in the presence of methicillin.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Beck W. D., Berger-Bächi B., Kayser F. H. Additional DNA in methicillin-resistant Staphylococcus aureus and molecular cloning of mec-specific DNA. J Bacteriol. 1986 Feb;165(2):373–378. doi: 10.1128/jb.165.2.373-378.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger-Bächi B., Barberis-Maino L., Strässle A., Kayser F. H. FemA, a host-mediated factor essential for methicillin resistance in Staphylococcus aureus: molecular cloning and characterization. Mol Gen Genet. 1989 Oct;219(1-2):263–269. doi: 10.1007/BF00261186. [DOI] [PubMed] [Google Scholar]
- Blümel P., Uecker W., Giesbrecht P. Zero order kinetics of cell wall turnover in Staphylococcus aureus. Arch Microbiol. 1979 May;121(2):103–110. doi: 10.1007/BF00689972. [DOI] [PubMed] [Google Scholar]
- Gustafson J. E., Wilkinson B. J. Lower autolytic activity in a homogeneous methicillin-resistant Staphylococcus aureus strain compared to derived heterogeneous-resistant and susceptible strains. FEMS Microbiol Lett. 1989 May;50(1-2):107–111. doi: 10.1016/0378-1097(89)90468-0. [DOI] [PubMed] [Google Scholar]
- Hartman B. J., Tomasz A. Expression of methicillin resistance in heterogeneous strains of Staphylococcus aureus. Antimicrob Agents Chemother. 1986 Jan;29(1):85–92. doi: 10.1128/aac.29.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman B. J., Tomasz A. Low-affinity penicillin-binding protein associated with beta-lactam resistance in Staphylococcus aureus. J Bacteriol. 1984 May;158(2):513–516. doi: 10.1128/jb.158.2.513-516.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornblum J., Hartman B. J., Novick R. P., Tomasz A. Conversion of a homogeneously methicillin-resistant strain of Staphylococcus aureus to heterogeneous resistance by Tn551-mediated insertional inactivation. Eur J Clin Microbiol. 1986 Dec;5(6):714–718. doi: 10.1007/BF02013311. [DOI] [PubMed] [Google Scholar]
- Kuhl S. A., Pattee P. A., Baldwin J. N. Chromosomal map location of the methicillin resistance determinant in Staphylococcus aureus. J Bacteriol. 1978 Aug;135(2):460–465. doi: 10.1128/jb.135.2.460-465.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuhashi M., Song M. D., Ishino F., Wachi M., Doi M., Inoue M., Ubukata K., Yamashita N., Konno M. Molecular cloning of the gene of a penicillin-binding protein supposed to cause high resistance to beta-lactam antibiotics in Staphylococcus aureus. J Bacteriol. 1986 Sep;167(3):975–980. doi: 10.1128/jb.167.3.975-980.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews P. R., Reed K. C., Stewart P. R. The cloning of chromosomal DNA associated with methicillin and other resistances in Staphylococcus aureus. J Gen Microbiol. 1987 Jul;133(7):1919–1929. doi: 10.1099/00221287-133-7-1919. [DOI] [PubMed] [Google Scholar]
- Murakami K., Tomasz A. Involvement of multiple genetic determinants in high-level methicillin resistance in Staphylococcus aureus. J Bacteriol. 1989 Feb;171(2):874–879. doi: 10.1128/jb.171.2.874-879.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qoronfleh M. W., Wilkinson B. J. Effects of growth of methicillin-resistant and -susceptible Staphylococcus aureus in the presence of beta-lactams on peptidoglycan structure and susceptibility to lytic enzymes. Antimicrob Agents Chemother. 1986 Feb;29(2):250–257. doi: 10.1128/aac.29.2.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raychaudhuri D., Chatterjee A. N. Use of resistant mutants to study the interaction of triton X-100 with Staphylococcus aureus. J Bacteriol. 1985 Dec;164(3):1337–1349. doi: 10.1128/jb.164.3.1337-1349.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TOMASZ A., JAMIESON J. D., OTTOLENGHI E. THE FINE STRUCTURE OF DIPLOCOCCUS PNEUMONIAE. J Cell Biol. 1964 Aug;22:453–467. doi: 10.1083/jcb.22.2.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson N. E., Pattee P. A. Transformation in Staphylococcus aureus: role of bacteriophage and incidence of competence among strains. J Bacteriol. 1977 Feb;129(2):778–788. doi: 10.1128/jb.129.2.778-788.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomasz A., Drugeon H. B., de Lencastre H. M., Jabes D., McDougall L., Bille J. New mechanism for methicillin resistance in Staphylococcus aureus: clinical isolates that lack the PBP 2a gene and contain normal penicillin-binding proteins with modified penicillin-binding capacity. Antimicrob Agents Chemother. 1989 Nov;33(11):1869–1874. doi: 10.1128/aac.33.11.1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utsui Y., Yokota T. Role of an altered penicillin-binding protein in methicillin- and cephem-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 1985 Sep;28(3):397–403. doi: 10.1128/aac.28.3.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson R., Tomasz A. Inhibition of cell wall synthesis and acylation of the penicillin binding proteins during prolonged exposure of growing Streptococcus pneumoniae to benzylpenicillin. Eur J Biochem. 1985 Sep 16;151(3):475–483. doi: 10.1111/j.1432-1033.1985.tb09126.x. [DOI] [PubMed] [Google Scholar]