Abstract
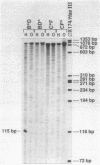
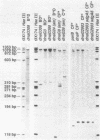
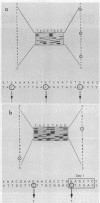
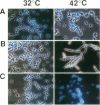
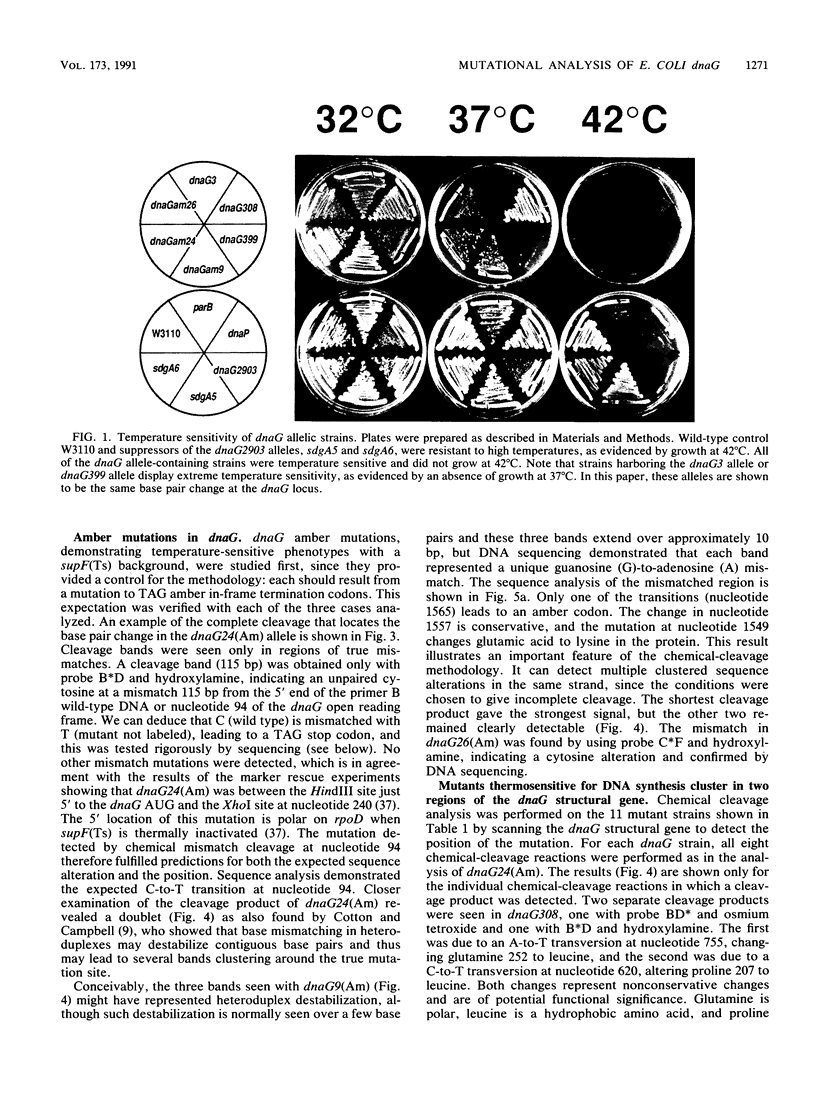
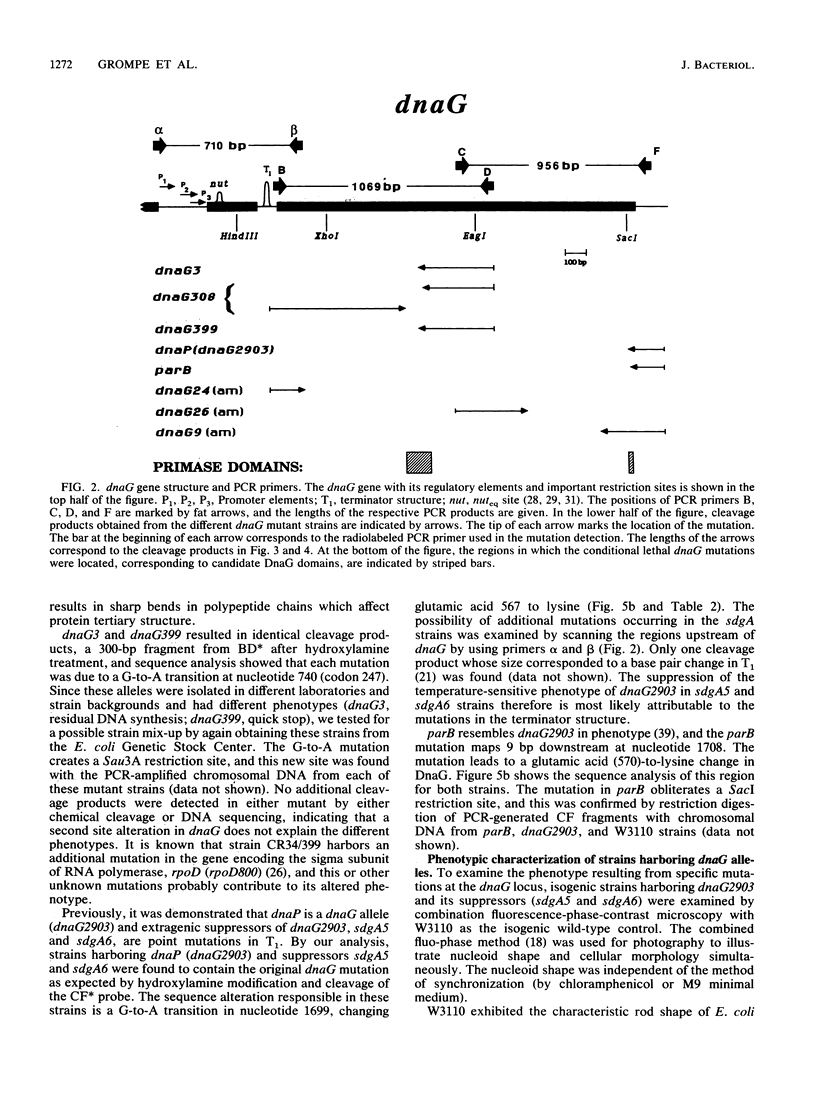
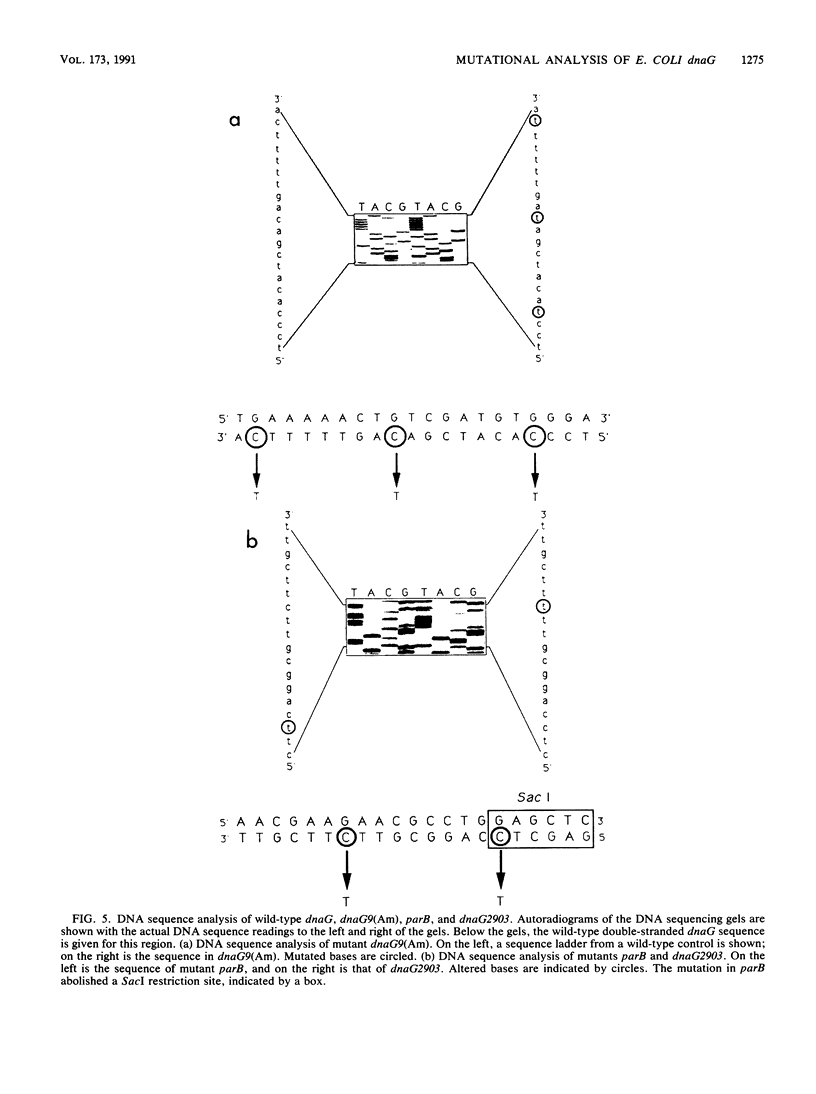
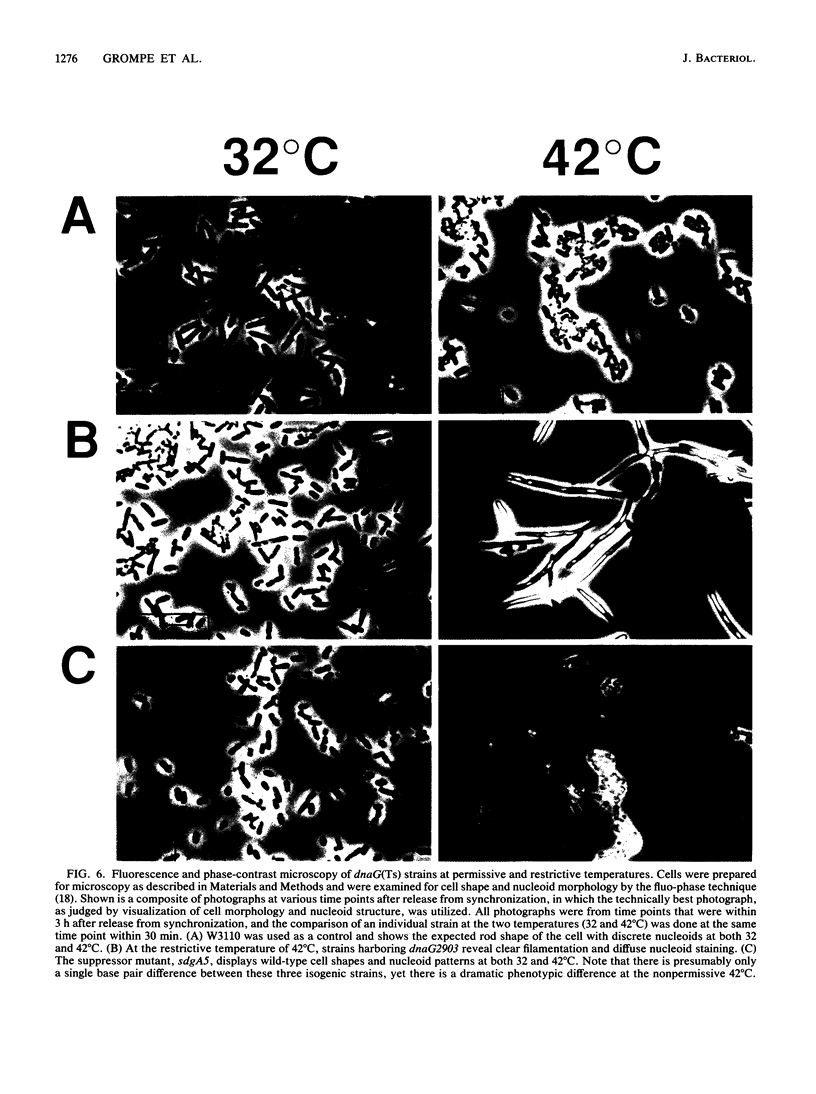
Eleven conditional lethal dnaG(Ts) mutations were located by chemical cleavage of heteroduplexes formed between polymerase chain reaction-amplified DNAs from wild-type and mutant dnaG genes. This entailed end labeling one DNA strand of the heteroduplex, chemically modifying the strands with hydroxylamine or osmium tetroxide (OsO4) at the site of mismatch, and cleaving them with piperidine. The cleavage products were electrophoresed, and the size corresponded to the position of the mutation with respect to the labeled primer. Exact base pair changes were then determined by DNA sequence analysis. The dnaG3, dnaG308, and dnaG399 mutations map within 135 nucleotides of one another near the middle of dnaG. The "parB" allele of dnaG is 36 bp from the 3' end of dnaG and 9 bp downstream of dnaG2903; both appear to result in abnormal chromosome partitioning and diffuse nucleoid staining. A suppressor of the dnaG2903 allele (sdgA5) maps within the terminator T1 just 5' to the dnaG gene. Isogenic strains that carried dnaG2903 and did or did not carry the sdgA5 suppressor were analyzed by a combination of phase-contrast and fluorescence microscopy with 4',6-diamidino-2-phenylindole to stain DNA and visualize the partitioning chromosome. Overexpression of the mutant dnaG allele corrected the abnormal diffuse-nucleoid-staining phenotype associated with normally expressed dnaG2903. The mutations within the dnaG gene appear to cluster into two regions which may represent distinct functional domains within the primase protein.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Almond N., Yajnik V., Svec P., Godson G. N. An Escherichia coli cis-acting antiterminator sequence: the dnaG nut site. Mol Gen Genet. 1989 Apr;216(2-3):195–203. doi: 10.1007/BF00334356. [DOI] [PubMed] [Google Scholar]
- BONHOEFFER F., SCHALLER H. A METHOD FOR SELECTIVE ENRICHMENT OF MUTANTS BASED ON THE HIGH UV SENSITIVITY OF DNA CONTAINING 5-BROMOURACIL. Biochem Biophys Res Commun. 1965 Jun 18;20:93–97. [PubMed] [Google Scholar]
- Bouché J. P., Rowen L., Kornberg A. The RNA primer synthesized by primase to initiate phage G4 DNA replication. J Biol Chem. 1978 Feb 10;253(3):765–769. [PubMed] [Google Scholar]
- Bouché J. P., Zechel K., Kornberg A. dnaG gene product, a rifampicin-resistant RNA polymerase, initiates the conversion of a single-stranded coliphage DNA to its duplex replicative form. J Biol Chem. 1975 Aug 10;250(15):5995–6001. [PubMed] [Google Scholar]
- Boyer H. W., Roulland-Dussoix D. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J Mol Biol. 1969 May 14;41(3):459–472. doi: 10.1016/0022-2836(69)90288-5. [DOI] [PubMed] [Google Scholar]
- Burton Z. F., Gross C. A., Watanabe K. K., Burgess R. R. The operon that encodes the sigma subunit of RNA polymerase also encodes ribosomal protein S21 and DNA primase in E. coli K12. Cell. 1983 Feb;32(2):335–349. doi: 10.1016/0092-8674(83)90453-1. [DOI] [PubMed] [Google Scholar]
- Carl P. L. Escherichia coli mutants with temperature-sensitive synthesis of DNA. Mol Gen Genet. 1970;109(2):107–122. doi: 10.1007/BF00269647. [DOI] [PubMed] [Google Scholar]
- Cotton R. G., Campbell R. D. Chemical reactivity of matched cytosine and thymine bases near mismatched and unmatched bases in a heteroduplex between DNA strands with multiple differences. Nucleic Acids Res. 1989 Jun 12;17(11):4223–4233. doi: 10.1093/nar/17.11.4223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotton R. G. Detection of single base changes in nucleic acids. Biochem J. 1989 Oct 1;263(1):1–10. doi: 10.1042/bj2630001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotton R. G., Rodrigues N. R., Campbell R. D. Reactivity of cytosine and thymine in single-base-pair mismatches with hydroxylamine and osmium tetroxide and its application to the study of mutations. Proc Natl Acad Sci U S A. 1988 Jun;85(12):4397–4401. doi: 10.1073/pnas.85.12.4397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahl H. H., Lamande S. R., Cotton R. G., Bateman J. F. Detection and localization of base changes in RNA using a chemical cleavage method. Anal Biochem. 1989 Dec;183(2):263–268. doi: 10.1016/0003-2697(89)90477-6. [DOI] [PubMed] [Google Scholar]
- Erickson B. D., Burton Z. F., Watanabe K. K., Burgess R. R. Nucleotide sequence of the rpsU-dnaG-rpoD operon from Salmonella typhimurium and a comparison of this sequence with the homologous operon of Escherichia coli. Gene. 1985;40(1):67–78. doi: 10.1016/0378-1119(85)90025-3. [DOI] [PubMed] [Google Scholar]
- Grompe M., Muzny D. M., Caskey C. T. Scanning detection of mutations in human ornithine transcarbamoylase by chemical mismatch cleavage. Proc Natl Acad Sci U S A. 1989 Aug;86(15):5888–5892. doi: 10.1073/pnas.86.15.5888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross J. D. DNA replication in bacteria. Curr Top Microbiol Immunol. 1972;57:39–74. doi: 10.1007/978-3-642-65297-4_2. [DOI] [PubMed] [Google Scholar]
- Hiasa H., Sakai H., Komano T., Godson G. N. Structural features of the priming signal recognized by primase: mutational analysis of the phage G4 origin of complementary DNA strand synthesis. Nucleic Acids Res. 1990 Aug 25;18(16):4825–4831. doi: 10.1093/nar/18.16.4825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiasa H., Sakai H., Tanaka K., Honda Y., Komano T., Godson G. N. Mutational analysis of the primer RNA template region in the replication origin (oric) of bacteriophage G4: priming signal recognition by Escherichia coli primase. Gene. 1989 Dec 7;84(1):9–16. doi: 10.1016/0378-1119(89)90133-9. [DOI] [PubMed] [Google Scholar]
- Hiasa H., Tanaka K., Sakai H., Yoshida K., Honda Y., Komano T., Godson G. N. Distinct functional contributions of three potential secondary structures in the phage G4 origin of complementary DNA strand synthesis. Gene. 1989 Dec 7;84(1):17–22. doi: 10.1016/0378-1119(89)90134-0. [DOI] [PubMed] [Google Scholar]
- Hirota Y., Mordoh J., Jacob F. On the process of cellular division in Escherichia coli. 3. Thermosensitive mutants of Escherichia coli altered in the process of DNA initiation. J Mol Biol. 1970 Nov 14;53(3):369–387. doi: 10.1016/0022-2836(70)90072-0. [DOI] [PubMed] [Google Scholar]
- Hussain K., Elliott E. J., Salmond G. P. The parD- mutant of Escherichia coli also carries a gyrAam mutation. The complete sequence of gyrA. Mol Microbiol. 1987 Nov;1(3):259–273. doi: 10.1111/j.1365-2958.1987.tb01932.x. [DOI] [PubMed] [Google Scholar]
- Katayama T., Murakami Y., Wada C., Ohmori H., Yura T., Nagata T. Genetic suppression of a dnaG mutation in Escherichia coli. J Bacteriol. 1989 Mar;171(3):1485–1491. doi: 10.1128/jb.171.3.1485-1491.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato J., Nishimura Y., Imamura R., Niki H., Hiraga S., Suzuki H. New topoisomerase essential for chromosome segregation in E. coli. Cell. 1990 Oct 19;63(2):393–404. doi: 10.1016/0092-8674(90)90172-b. [DOI] [PubMed] [Google Scholar]
- Kato J., Nishimura Y., Suzuki H. Escherichia coli parA is an allele of the gyrB gene. Mol Gen Genet. 1989 May;217(1):178–181. doi: 10.1007/BF00330959. [DOI] [PubMed] [Google Scholar]
- Kogan S. C., Doherty M., Gitschier J. An improved method for prenatal diagnosis of genetic diseases by analysis of amplified DNA sequences. Application to hemophilia A. N Engl J Med. 1987 Oct 15;317(16):985–990. doi: 10.1056/NEJM198710153171603. [DOI] [PubMed] [Google Scholar]
- Lark K. G. Genetic control over the initiation of the synthesis of the short deoxynucleotide chains in E. coli. Nat New Biol. 1972 Dec 20;240(103):237–240. doi: 10.1038/newbio240237a0. [DOI] [PubMed] [Google Scholar]
- Liebke H., Gross C., Walter W., Burgess R. A new mutation rpoD800, affecting the sigma subunit of E. coli RNA polymerase is allelic to two other sigma mutants. Mol Gen Genet. 1980 Jan;177(2):277–282. doi: 10.1007/BF00267439. [DOI] [PubMed] [Google Scholar]
- Lupski J. R., Godson G. N. DNA----DNA, and DNA----RNA----protein: orchestration by a single complex operon. Bioessays. 1989 May;10(5):152–157. doi: 10.1002/bies.950100504. [DOI] [PubMed] [Google Scholar]
- Lupski J. R., Godson G. N. The rpsU-dnaG-rpoD macromolecular synthesis operon of E. coli. Cell. 1984 Dec;39(2 Pt 1):251–252. doi: 10.1016/0092-8674(84)90001-1. [DOI] [PubMed] [Google Scholar]
- Lupski J. R., Ruiz A. A., Godson G. N. Promotion, termination, and anti-termination in the rpsU-dnaG-rpoD macromolecular synthesis operon of E. coli K-12. Mol Gen Genet. 1984;195(3):391–401. doi: 10.1007/BF00341439. [DOI] [PubMed] [Google Scholar]
- Lupski J. R., Smiley B. L., Blattner F. R., Godson G. N. Cloning and characterization of the Escherichia coli chromosomal region surrounding the dnaG Gene, with a correlated physical and genetic map of dnaG generated via transposon Tn5 mutagenesis. Mol Gen Genet. 1982;185(1):120–128. doi: 10.1007/BF00333800. [DOI] [PubMed] [Google Scholar]
- Lupski J. R., Smiley B. L., Godson G. N. Regulation of the rpsU-dnaG-rpoD macromolecular synthesis operon and the initiation of DNA replication in Escherichia coli K-12. Mol Gen Genet. 1983;189(1):48–57. doi: 10.1007/BF00326054. [DOI] [PubMed] [Google Scholar]
- Marinus M. G., Adelberg E. A. Vegetative Replication and Transfer Replication of Deoxyribonucleic Acid in Temperature-Sensitive Mutants of Escherichia coli K-12. J Bacteriol. 1970 Dec;104(3):1266–1272. doi: 10.1128/jb.104.3.1266-1272.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurer R., Osmond B. C., Shekhtman E., Wong A., Botstein D. Functional interchangeability of DNA replication genes in Salmonella typhimurium and Escherichia coli demonstrated by a general complementation procedure. Genetics. 1984 Sep;108(1):1–23. doi: 10.1093/genetics/108.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mead D. A., Szczesna-Skorupa E., Kemper B. Single-stranded DNA 'blue' T7 promoter plasmids: a versatile tandem promoter system for cloning and protein engineering. Protein Eng. 1986 Oct-Nov;1(1):67–74. doi: 10.1093/protein/1.1.67. [DOI] [PubMed] [Google Scholar]
- Murakami Y., Nagata T., Schwarz W., Wada C., Yura T. Novel dnaG mutation in a dnaP mutant of Escherichia coli. J Bacteriol. 1985 May;162(2):830–832. doi: 10.1128/jb.162.2.830-832.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura Y. Amber dnaG mutation exerting a polar effect on the synthesis of RNA polymerase sigma factor in Escherichia coli. Mol Gen Genet. 1984;196(1):179–182. doi: 10.1007/BF00334114. [DOI] [PubMed] [Google Scholar]
- Nesin M., Lupski J. R., Godson G. N. Role of the 5' upstream sequence and tandem promoters in regulation of the rpsU-dnaG-rpoD macromolecular synthesis operon. J Bacteriol. 1988 Dec;170(12):5759–5764. doi: 10.1128/jb.170.12.5759-5764.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norris V., Alliotte T., Jaffé A., D'Ari R. DNA replication termination in Escherichia coli parB (a dnaG allele), parA, and gyrB mutants affected in DNA distribution. J Bacteriol. 1986 Nov;168(2):494–504. doi: 10.1128/jb.168.2.494-504.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunn W. D., Tropp B. E. Effects of phenethyl alcohol on phospholipid metabolism in Escherichia coli. J Bacteriol. 1972 Jan;109(1):162–168. doi: 10.1128/jb.109.1.162-168.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Päbo S., Wilson A. C. Polymerase chain reaction reveals cloning artefacts. Nature. 1988 Aug 4;334(6181):387–388. doi: 10.1038/334387b0. [DOI] [PubMed] [Google Scholar]
- Rowen L., Kornberg A. A ribo-deoxyribonucleotide primer synthesized by primase. J Biol Chem. 1978 Feb 10;253(3):770–774. [PubMed] [Google Scholar]
- Rowen L., Kornberg A. Primase, the dnaG protein of Escherichia coli. An enzyme which starts DNA chains. J Biol Chem. 1978 Feb 10;253(3):758–764. [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver S., Wendt L. Mechanism of action of phenethyl alcohol: breakdown of the cellular permeability barrier. J Bacteriol. 1967 Feb;93(2):560–566. doi: 10.1128/jb.93.2.560-566.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smiley B. L., Lupski J. R., Svec P. S., McMacken R., Godson G. N. Sequences of the Escherichia coli dnaG primase gene and regulation of its expression. Proc Natl Acad Sci U S A. 1982 Aug;79(15):4550–4554. doi: 10.1073/pnas.79.15.4550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada C., Yura T. Phenethyl alcohol resistance in Escherichia coli. 3. A temperature-sensitive mutation(dnaP) affecting DNA replication. Genetics. 1974 Jun;77(2):199–220. doi: 10.1093/genetics/77.2.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L. F., Doi R. H. Nucleotide sequence and organization of Bacillus subtilis RNA polymerase major sigma (sigma 43) operon. Nucleic Acids Res. 1986 May 27;14(10):4293–4307. doi: 10.1093/nar/14.10.4293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wechsler J. A., Gross J. D. Escherichia coli mutants temperature-sensitive for DNA synthesis. Mol Gen Genet. 1971;113(3):273–284. doi: 10.1007/BF00339547. [DOI] [PubMed] [Google Scholar]
- Wold M. S., McMacken R. Regulation of expression of the Escherichia coli dnaG gene and amplification of the dnaG primase. Proc Natl Acad Sci U S A. 1982 Aug;79(16):4907–4911. doi: 10.1073/pnas.79.16.4907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Ende A., Baker T. A., Ogawa T., Kornberg A. Initiation of enzymatic replication at the origin of the Escherichia coli chromosome: primase as the sole priming enzyme. Proc Natl Acad Sci U S A. 1985 Jun;82(12):3954–3958. doi: 10.1073/pnas.82.12.3954. [DOI] [PMC free article] [PubMed] [Google Scholar]