Abstract
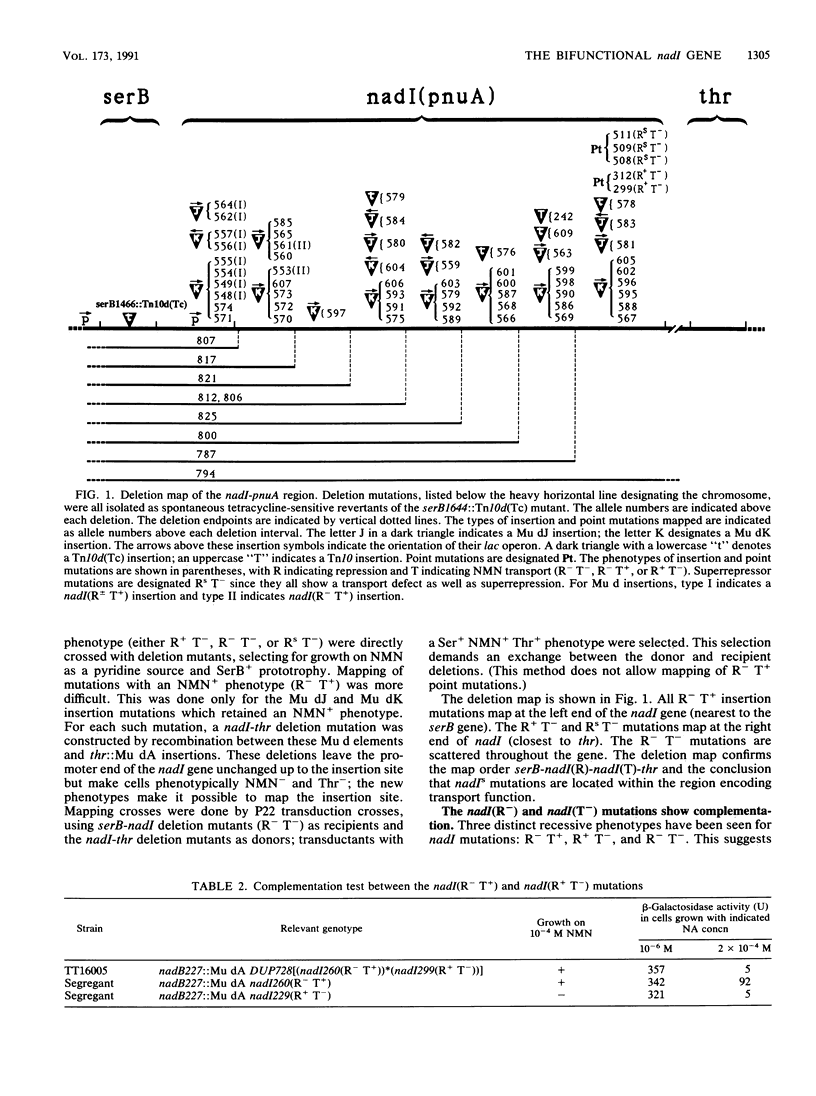
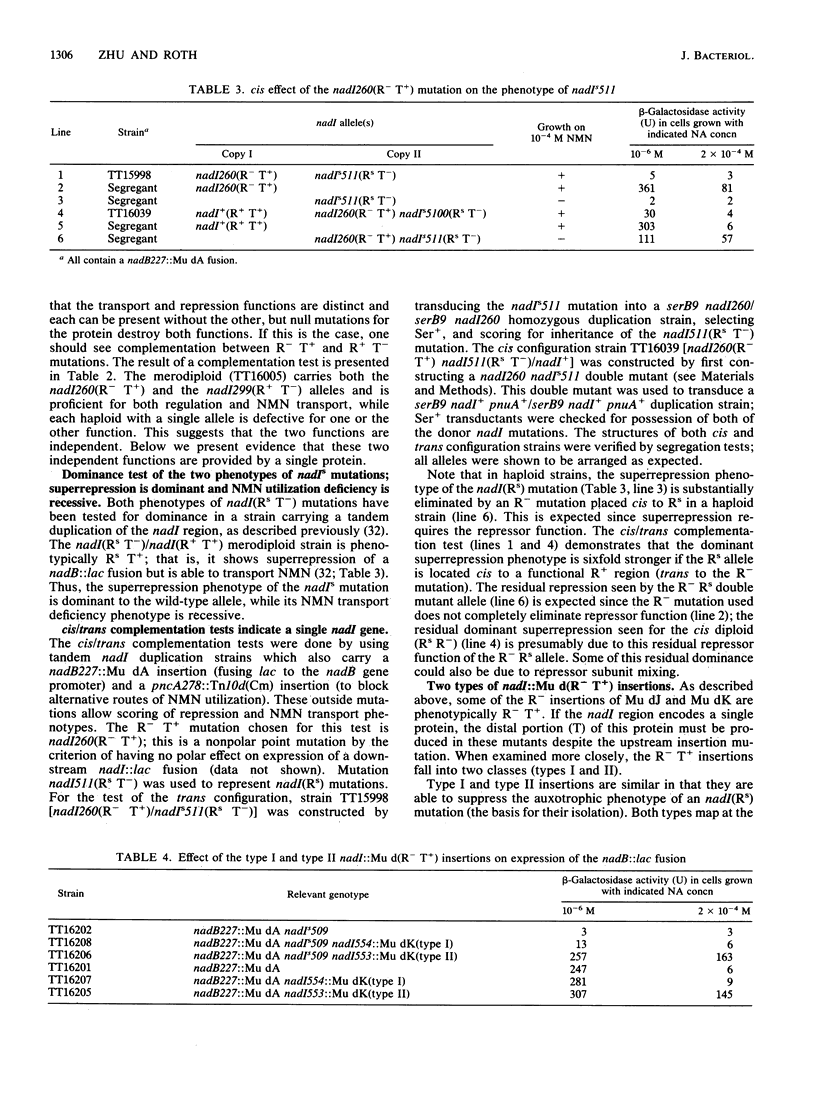
Mutants of the nadI and pnuA genes were independently isolated on the basis of defects in repression of NAD biosynthetic genes and defects in transport nicotinamide mononucleotide (NMN). The mutations map at min 99 on the Salmonella chromosome, and the affected regions appear to be cotranscribed. Some pairs of nadI and pnuA mutations complement, suggesting the existence of independent functions. However, cis/trans tests with particular mutations provide evidence that both repressor and transport functions are actually performed by a single bifunctional protein. (This result confirms sequencing data of Foster and coworkers [J. W. Foster, Y. K. Park, T. Fenger, and M. P. Spector, J. Bacteriol. 172:4187-4196, 1990]). We have designated the gene for this bifunctional protein nadI and distinguish the regulatory and transport defects with phenotypic designations (R and T). When a nadI(R- T+) mutation (eliminating only repression function) is placed cis to a superrepressor mutation, nadI(Rs T-), the superrepression phenotype is lost. In contrast, placement of R- and Rs T- mutations in trans allows full superrepression. This result suggests that the transport function (eliminated by the Rs T- mutation) and the repression function are provided by the same protein. Insertion mutations in the promoter-proximal repressor region of the nadI gene eliminate transport function unless the inserted element can provide both for both transcription and translation start signals; this finding suggests that there is no transcriptional or translational start between the regions encoding repression and transport functions.
Full text
PDF






Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Blazey D. L., Burns R. O. Transcriptional activity of the transposable element Tn10 in the Salmonella typhimurium ilvGEDA operon. Proc Natl Acad Sci U S A. 1982 Aug;79(16):5011–5015. doi: 10.1073/pnas.79.16.5011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochner B. R., Huang H. C., Schieven G. L., Ames B. N. Positive selection for loss of tetracycline resistance. J Bacteriol. 1980 Aug;143(2):926–933. doi: 10.1128/jb.143.2.926-933.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brendel V., Trifonov E. N. A computer algorithm for testing potential prokaryotic terminators. Nucleic Acids Res. 1984 May 25;12(10):4411–4427. doi: 10.1093/nar/12.10.4411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadaban M. J., Cohen S. N. Lactose genes fused to exogenous promoters in one step using a Mu-lac bacteriophage: in vivo probe for transcriptional control sequences. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4530–4533. doi: 10.1073/pnas.76.9.4530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castilho B. A., Olfson P., Casadaban M. J. Plasmid insertion mutagenesis and lac gene fusion with mini-mu bacteriophage transposons. J Bacteriol. 1984 May;158(2):488–495. doi: 10.1128/jb.158.2.488-495.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan R. K., Botstein D., Watanabe T., Ogata Y. Specialized transduction of tetracycline resistance by phage P22 in Salmonella typhimurium. II. Properties of a high-frequency-transducing lysate. Virology. 1972 Dec;50(3):883–898. doi: 10.1016/0042-6822(72)90442-4. [DOI] [PubMed] [Google Scholar]
- Chumley F. G., Menzel R., Roth J. R. Hfr formation directed by tn10. Genetics. 1979 Apr;91(4):639–655. doi: 10.1093/genetics/91.4.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciampi M. S., Roth J. R. Polarity effects in the hisG gene of salmonella require a site within the coding sequence. Genetics. 1988 Feb;118(2):193–202. doi: 10.1093/genetics/118.2.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciampi M. S., Schmid M. B., Roth J. R. Transposon Tn10 provides a promoter for transcription of adjacent sequences. Proc Natl Acad Sci U S A. 1982 Aug;79(16):5016–5020. doi: 10.1073/pnas.79.16.5016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cookson B. T., Olivera B. M., Roth J. R. Genetic characterization and regulation of the nadB locus of Salmonella typhimurium. J Bacteriol. 1987 Sep;169(9):4285–4293. doi: 10.1128/jb.169.9.4285-4293.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demerec M., Adelberg E. A., Clark A. J., Hartman P. E. A proposal for a uniform nomenclature in bacterial genetics. Genetics. 1966 Jul;54(1):61–76. doi: 10.1093/genetics/54.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott T., Roth J. R. Characterization of Tn10d-Cam: a transposition-defective Tn10 specifying chloramphenicol resistance. Mol Gen Genet. 1988 Aug;213(2-3):332–338. doi: 10.1007/BF00339599. [DOI] [PubMed] [Google Scholar]
- Foster J. W., Holley-Guthrie E. A., Warren F. Regulation of NAD metabolism in Salmonella typhimurium: genetic analysis and cloning of the nadR repressor locus. Mol Gen Genet. 1987 Jun;208(1-2):279–287. doi: 10.1007/BF00330454. [DOI] [PubMed] [Google Scholar]
- Foster J. W., Kinney D. M., Moat A. G. Pyridine nucleotide cycle of Salmonella typhimurium: isolation and characterization of pncA, pncB, and pncC mutants and utilization of exogenous nicotinamide adenine dinucleotide. J Bacteriol. 1979 Mar;137(3):1165–1175. doi: 10.1128/jb.137.3.1165-1175.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster J. W., Park Y. K., Penfound T., Fenger T., Spector M. P. Regulation of NAD metabolism in Salmonella typhimurium: molecular sequence analysis of the bifunctional nadR regulator and the nadA-pnuC operon. J Bacteriol. 1990 Aug;172(8):4187–4196. doi: 10.1128/jb.172.8.4187-4196.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goosen N., van de Putte P. Role of ner protein in bacteriophage Mu transposition. J Bacteriol. 1986 Aug;167(2):503–507. doi: 10.1128/jb.167.2.503-507.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holley E. A., Spector M. P., Foster J. W. Regulation of NAD biosynthesis in Salmonella typhimurium: expression of nad-lac gene fusions and identification of a nad regulatory locus. J Gen Microbiol. 1985 Oct;131(10):2759–2770. doi: 10.1099/00221287-131-10-2759. [DOI] [PubMed] [Google Scholar]
- Hughes K. T., Roth J. R. Conditionally transposition-defective derivative of Mu d1(Amp Lac). J Bacteriol. 1984 Jul;159(1):130–137. doi: 10.1128/jb.159.1.130-137.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes K. T., Roth J. R. Transitory cis complementation: a method for providing transposition functions to defective transposons. Genetics. 1988 May;119(1):9–12. doi: 10.1093/genetics/119.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinney D. M., Foster J. W., Moat A. G. Pyridine nucleotide cycle of Salmonella typhimurium: in vitro demonstration of nicotinamide mononucleotide deamidase and characterization of pnuA mutants defective in nicotinamide mononucleotide transport. J Bacteriol. 1979 Nov;140(2):607–611. doi: 10.1128/jb.140.2.607-611.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maloy S. R., Nunn W. D. Selection for loss of tetracycline resistance by Escherichia coli. J Bacteriol. 1981 Feb;145(2):1110–1111. doi: 10.1128/jb.145.2.1110-1111.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maloy S. R., Roth J. R. Regulation of proline utilization in Salmonella typhimurium: characterization of put::Mu d(Ap, lac) operon fusions. J Bacteriol. 1983 May;154(2):561–568. doi: 10.1128/jb.154.2.561-568.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuwald A. F., Stauffer G. V. DNA sequence and characterization of the Escherichia coli serB gene. Nucleic Acids Res. 1985 Oct 11;13(19):7025–7039. doi: 10.1093/nar/13.19.7025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid M. B., Roth J. R. Genetic methods for analysis and manipulation of inversion mutations in bacteria. Genetics. 1983 Nov;105(3):517–537. doi: 10.1093/genetics/105.3.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmieger H. A method for detection of phage mutants with altered transducing ability. Mol Gen Genet. 1971;110(4):378–381. doi: 10.1007/BF00438281. [DOI] [PubMed] [Google Scholar]
- Simons R. W., Hoopes B. C., McClure W. R., Kleckner N. Three promoters near the termini of IS10: pIN, pOUT, and pIII. Cell. 1983 Sep;34(2):673–682. doi: 10.1016/0092-8674(83)90400-2. [DOI] [PubMed] [Google Scholar]
- Spector M. P., Hill J. M., Holley E. A., Foster J. W. Genetic characterization of pyridine nucleotide uptake mutants of Salmonella typhimurium. J Gen Microbiol. 1985 Jun;131(6):1313–1322. doi: 10.1099/00221287-131-6-1313. [DOI] [PubMed] [Google Scholar]
- VOGEL H. J., BONNER D. M. Acetylornithinase of Escherichia coli: partial purification and some properties. J Biol Chem. 1956 Jan;218(1):97–106. [PubMed] [Google Scholar]
- Way J. C., Davis M. A., Morisato D., Roberts D. E., Kleckner N. New Tn10 derivatives for transposon mutagenesis and for construction of lacZ operon fusions by transposition. Gene. 1984 Dec;32(3):369–379. doi: 10.1016/0378-1119(84)90012-x. [DOI] [PubMed] [Google Scholar]
- Zhu N., Olivera B. M., Roth J. R. Activity of the nicotinamide mononucleotide transport system is regulated in Salmonella typhimurium. J Bacteriol. 1991 Feb;173(3):1311–1320. doi: 10.1128/jb.173.3.1311-1320.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N., Olivera B. M., Roth J. R. Genetic characterization of the pnuC gene, which encodes a component of the nicotinamide mononucleotide transport system in Salmonella typhimurium. J Bacteriol. 1989 Aug;171(8):4402–4409. doi: 10.1128/jb.171.8.4402-4409.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N., Olivera B. M., Roth J. R. Identification of a repressor gene involved in the regulation of NAD de novo biosynthesis in Salmonella typhimurium. J Bacteriol. 1988 Jan;170(1):117–125. doi: 10.1128/jb.170.1.117-125.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]



