Abstract
The glutamate receptor subunit B (GluR-B) pre-mRNA is edited at two adenosine residues, resulting in amino acid changes that alter the electrophysiologic properties of the glutamate receptor. Previous studies showed that these amino acid changes are due to adenosine to inosine conversions in two codons resulting from adenosine deamination. Here, we describe the purification and characterization of an activity from human HeLa cells that efficiently and accurately edits GluR-B pre-mRNA at both of these sites. The purified activity contains a human homolog of the recently reported rat RED1 (rRED1) protein, a member of the family of double-stranded RNA-dependent deaminase proteins. Recombinant human RED1 (hRED1), but not recombinant dsRAD, another member of the family, efficiently edits both the Q/R and R/G sites of GluR-B RNA. We conclude that the GluR-B editing activity present in HeLa cell extracts and the recombinant hRED1 protein are indistinguishable.
Keywords: RNA editing, inosine, ion channel
RNA editing is a novel mechanism for generating distinct protein isoforms from a single pre-mRNA (1–3). Pre-mRNA encoding the B subunit of the glutamate receptor (GluR-B) undergoes site specific deamination (4–6) at an adenosine residue in the second transmembrane domain. This results in the transformation of a glutamate codon (CAG) to an arginine codon (CIG) (designated the Q/R site) (7–11). This amino acid substitution profoundly alters the ion permeability and conductance properties of the class of α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid-responsive receptor channels in the brain that contain a GluR-B subunit (1, 12–13). More than 99% of the GluR-B mRNA is edited at the Q/R site in adult neurons (8–9). Adenosine conversions are also observed at the same position of the high-affinity kainate receptor subunits GluR-5 and GluR-6 with efficiencies of 40% and 80%, respectively (8). In addition to the Q/R site, RNA adenosine deamination occurs in exon 13 of GluR-B, C, and D pre-mRNAs, where an arginine codon (AGA) is converted to a glycine codon (IGA) (designated the R/G site) (14–15).
Editing of both the Q/R and R/G site requires base pairing between the editing site and an editing site complementary sequence (ECS) within the downstream intron (10). The requirement for RNA duplex formation at the editing site in GluR-B was demonstrated by showing that base substitutions that disrupt the potential for base pairing interactions block editing. Moreover, these mutants can be rescued in vivo and in vitro by second site mutations that reestablish base pairing (4, 10). These observations suggested that a double-stranded (ds)RNA adenosine deaminase activity may be involved in GluR-B editing. Two dsRNA-dependent adenosine deaminases have been described in vertebrates: dsRAD and RED1. In vitro, recombinant RED1 accurately and efficiently edits GluR-B at the Q/R site (16). In contrast, recombinant dsRAD is capable of only low levels of GluR-B editing at the Q/R site (17–18), but converts A to I at sites that are not observed in vivo. These experiments with purified recombinant proteins suggest that RED1 is responsible for editing GluR-B RNA in vivo.
We have previously developed an in vitro system in which extracts of HeLa cells accurately and efficiently edit the Q/R site of GluR-B (4). In this study, we provide independent evidence that the GluR-B editing activity is indeed mediated by RED1. We have fractionated HeLa extracts into two dsRNA-dependent adenosine deaminase activities. One activity, identified as human dsRAD, converts A to I in dsRNA nonspecifically and lacks specific GluR-B editing activity at the Q/R site. A second deaminase activity, identified as hRED1, specifically edits GluR-B RNA at the Q/R and R/G sites. Moreover, we have expressed the cloned hRED1 in a baculovirus expression system, and we demonstrate that this enzyme efficiently edits GluR-B pre-mRNA at both the Q/R and R/G sites, and is thus indistinguishable from the activity in HeLa cell nuclear extracts. The recombinant RED1 was also used to characterize the RNA sequence requirements for editing at the R/G site. Our results suggest that editing at both the Q/R and R/G sites could result from the activity of a single enzyme, RED1.
MATERIALS AND METHODS
Materials.
HeLa cells were purchased from the National Cell Culture Center (Minneapolis). Heparin Sepharose, Q Sepharose, phenyl-superose, and Mono Q HR 5/5 columns were purchased from Pharmacia. RNase P1 and adenylyl guanosine dinucleotide (ApG) were purchased from Sigma. Protein quantitation was done with a Bio-Rad protein assay kit. All oligonucleotides used for mutagenesis and cloning were made commercially (Operon Technologies, Alameda, CA; and Oligo Synthesis at Tufts, Boston).
Preparation of Nuclear Extract for Editing.
Nuclear extract for RNA editing was prepared as follows: A cell pellet from 100 l HeLa cells was quickly washed once and resuspended in wash buffer (10 mM Hepes, pH 7.9/1.5 mM MgCl2/10 mM KCl/0.5 mM dithiothreitol). Cells were lysed with a Dounce homogenizer, centrifuged at 3,000 × g for 15 min and the nuclear pellet was resuspended in one volume of low salt buffer (20 mM Tris⋅HCl/25% glycerol/1.5 mM MgCl2/15 mM KCl). One volume of high salt buffer (20 mM Tris⋅HCl/25% glycerol/1.5 mM MgCl2/1.2 M KCl) was then slowly added over 20 min and stirred slowly for 30 min. After centrifugation at 10,000 × g for 30 min, the supernatant was dialyzed against 10 volumes of editing buffer (20 mM Hepes, pH 8.0/100 mM KCl/5 mM dithiothreitol/10 mM EDTA/20% glycerol/0.5% Nonidet P-40). The nuclear extract was then precipitated with 20% saturated (NH4)2SO4 and the supernatant was precipitated with 45% saturated (NH4)2SO4. The 45% pellet then was resuspended in 100 ml of editing buffer and dialyzed against the same buffer. Aliquots were frozen at ⋅80°C. Editing was assayed as previously described using the 5′-end-labeled 3′-half GluR-B Q/R site RNA or the uniformly labeled ds α-tropomyosin exon 2 RNA (4).
Chromatography.
A 10-ml heparin Sepharose column (Pharmacia) was packed and equilibrated with buffer A (10 mM Hepes/50 mM KCl/5 mM EDTA/0.5 mM dithiothreitol). Twenty-five milliliters of HeLa nuclear extract was thawed, loaded onto the column and washed with the same buffer until OD280 reached baseline. The bound proteins were then eluted with a gradient of buffer B (buffer A + 1 M NaCl) to 100% in 30 ml, followed by a 30 ml wash with buffer B. The eluate fractions were collected and dialyzed individually against editing buffer. GluR-B RNA editing activity and dsRAD activity were independently assayed and the active fractions were pooled. These fractions were then chromatographed on a 5 ml Q Sepharose column followed by a 2 ml GluR-B RNA affinity column, as described for the heparin column. For the Q Sepharose column, the salt gradient was from 50 mM to 1M NaCl in 15 ml. For the GluR-B RNA affinity column, the gradient was from 50 mM to 3M KCl in 10 ml. Chromatography on the last hydrophobic column (5 ml of phenyl superose) was carried out by loading the active fractions in the high salt buffer [buffer A + 1 M NaCl + 0.5 M (NH4)2SO4] and eluting with decreasing concentrations of buffer A to 0% in 15 ml volume. All chromatography was done at 4°C. Active fractions were aliquoted and stored at −80°C.
Western Blot Analysis.
Recombinant His-tagged rRED1 was purified by nickel affinity chromatography according to the manufacturer’s protocol (Invitrogen) and used to raise antibodies in mice. Purified fractions from HeLa cells were separated on one- or two-dimensional gels, transferred to nitrocellulose membranes and blotted with antiserum (Bio-Rad).
Reverse Transcription-PCR, Cloning, and Sequencing.
Total RNA from HeLa cells was isolated using RNAzol (Tel-Test, Friendswood, TX) and mRNA was enriched using the PolyA Tract system (Promega). Reverse transcription and “nested” PCR reactions were performed using Vent DNA polymerase with degenerate primers (atg-n, ATGGATATNGANGANGANGANAANA; tga-n, TCANGGNGTNANNGANAANTG) and primers (up-n, GANAANATGAGTTCCAGCAGCA; down-n, GTCCTGCTCTGTGGGCTTCTCCACCCAGG). The amplified bands were purified from the agarose gel and cloned into pGEM7 at the SmaI site. To determine the sequence, the insert was deleted from the 5′ and 3′ ends using the Erase-a-Base system (Promega) and sequenced by dideoxy methods.
Expression of Recombinant Proteins in Insect Cells.
The human and rat RED1 cDNAs were subcloned into pFastBac-HTb and proteins were expressed in Sf9 cells (GIBCO/BRL). Proteins were purified by nickel column chromatography under native conditions (Invitrogen). The purified protein was >95% pure as determined by electrophoresis on an 8% SDS gel. One ml of the protein was dialyzed against PBS and the concentration was determined. The editing activities were determined as described (4). For the quick assay, the transfected cells were harvested, sonicated in editing buffer and centrifuged. The supernatant was then used without further purification to assay editing activity. Converted RNA was quantitated using the Fuji imaging system.
In Vitro Mutagenesis.
Mutagenesis in GluR-B RNA substrates was made by in vitro T7 transcription using PCR-generated DNA templates. RNA labeled specifically at the R/G site was made by RNA ligation as described (4).
UV Crosslinking.
The site specifically labeled GluR-B RNA (Q/R site) was mixed with the purified fraction and incubated for 5 min under editing conditions (4). The samples were put on ice and irradiated under 300 nm UV light for 10 min. The sample was resolved by SDS/PAGE (10% gel) and the crosslinked bands were visualized using the Fuji imaging system.
RESULTS
Separation of Two Distinct Adenosine Deaminase Activities from HeLa Cells.
We wished to identify the ds adenosine deaminase activity responsible for GluR-B editing in HeLa cell nuclear extracts. Two assays were employed to monitor adenosine deaminase activity, one which monitors site specific editing of GluR-B, and the other which detects nonspecific A to I conversion. In the first assay an RNA containing the Q/R site of GluR-B and the downstream ECS was labeled with 32P exclusively at the editing site as described (4). After incubation with the chromatographic fractions, the labeled RNA was digested with nuclease P1 and the products were separated by thin layer chromatography. Adenosine deaminase activity was demonstrated by the appearance of 32P-labeled inosine monophosphate. In the second assay, which detects nonspecific dsRNA editing, dsRNA was uniformly labeled with 32P, incubated with the sample and analyzed for A to I conversion as described above.
A 20–45% ammonium sulfate precipitate of the HeLa cell nuclear extract was fractionated on a heparin Sepharose affinity column and most of the dsRNA deaminase activity was recovered in the 0.5 M NaCl fractions, while the site-specific editing activity eluted at 0.9 M NaCl (Fig. 1A). When the fractions were analyzed by Western blot analysis using an anti-dsRAD antibody, a band of the expected molecular weight (140 kDa) was observed in the first peak (data not shown). The fractions in the second peak were pooled and chromatographed on a Q Sepharose column. The GluR-B RNA editing activity was found in a relatively sharp peak, whereas the dsRNA deaminase activity was present at low levels in the flow-through fractions and in fractions eluted at lower salt concentrations (Fig. 1B). The active fractions were then loaded onto a phenyl Sepharose hydrophobic interaction column and eluted with decreasing concentrations of NaCl. The editing activity was eluted at low concentrations of NaCl (Fig. 1C).
Figure 1.
Separation of two distinct adenosine deaminase activities from HeLa cell nuclear extracts. (A) Heparin affinity chromatography. (B) Q-Sepharose cation exchange chromatography. (C) Phenyl superose hydrophobic chromatography. The relative amount of editing activity in each fraction is indicated by the histogram at the bottom of the absorbency profile. Protein was monitored by absorbency at 280 nm (solid curves). Fractions were individually dialyzed before analyzing for GluR-B RNA editing (solid bar) and dsRNA deamination activities (open bar) by thin layer chromatography. Dashed lines indicate the salt gradient.
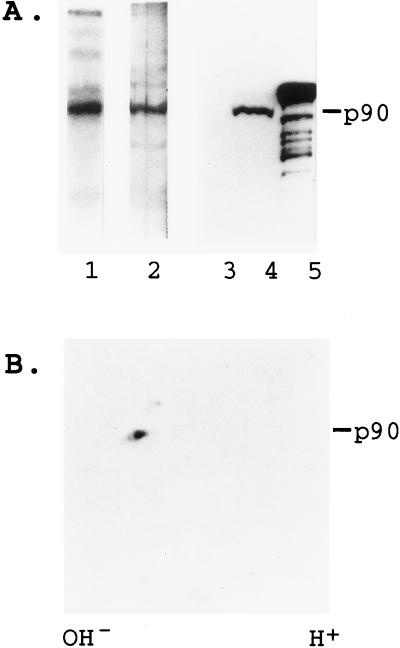
UV crosslinking experiments with the active phenyl Sepharose fractions and Q/R site-labeled GluR-B RNA revealed a strong band of 90 kDa and less intense bands of 140 and 100 kDa (Fig. 2A, lane 1). The 90-kDa protein could also be visualized by silver staining (Fig. 2A, lane 2). Western blot experiments using a polyclonal anti-rat RED1 antiserum (Fig. 2 A, lanes 3–5, and B) reveal a 90-kDa band demonstrating that RED1 was present in the active fractions. Thus the two adenosine deaminase activities can be chromatographically separated into one that specifically edits GluR-B and contains the 90-kDa protein RED1, and a second activity that nonspecifically deaminates dsRNA and contains the 140 kDa dsRAD.
Figure 2.
Identification of the protein required for GluR-B RNA editing in HeLa cells. (A) Lane 1: UV crosslinking of phenyl superose column fractions. GluR-B RNA labeled at the editing site was used in UV crosslinking experiments to detect proteins in the phenyl superose column fractions that bind to GluR-B RNA. The proteins were resolved by SDS/PAGE, and a 90-kDa crosslinked protein is indicated. Lane 2: Silver-stained SDS/PAGE of the most active fractions. Lane 3: Western blot of the flow-through. Lane 4: Western blot of the most active fraction. Lane 5: Western blot of recombinant rRED1 protein containing one ≈5 kDa His-tag at its N terminus. Western blot analysis was performed with anti-rat RED1 polyclonal antibodies. (B) The most active fraction was resolved on a two-dimensional gel [first dimension: isoelectric focusing pH 3–10; second dimension: SDS/PAGE (10% gel)] and by Western blot analysis with anti-rat RED1 polyclonal antibodies as above.
Characterization of Recombinant hRED1.
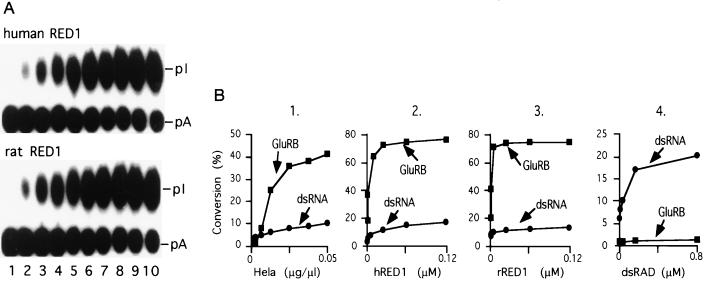
To characterize the properties of RED1 and compare them with the properties found in our HeLa cell extracts, we cloned the hRED1 cDNA from HeLa cell RNA by RT-PCR using two pairs of degenerate primers derived from the rRED1 sequence (10). One of the cloned cDNA sequence matches the Homo sapiens mRNA sequence deposited previously by Mittaz et al. in GenBank (accession no. X99383X99383) which is 86% homologous to rRED1 at the nucleic acid sequence level. Recombinant human and rat RED1 were synthesized and expressed in insect cells using the baculovirus expression system. A full-length protein of apparent molecular mass of 95 kDa was produced and purified on a nickel affinity column. The purified recombinant hRED1 edits GluR-B RNA efficiently at the Q/R site, and the kinetics of the reaction are identical to that of the rRED1 protein (Fig. 3A). Comparison of the purified HeLa cell activity and recombinant human and rat RED1 proteins reveals that the substrate specificities of the two deaminase activities are virtually indistinguishable (Fig. 3B). The ratio of the GluR-B editing and dsRNA deaminase activities is ≈80:1. By contrast, at concentrations of dsRAD at which maximal levels of activity with dsRNA are observed, no GluR-B editing activity is detected (Fig. 3B, panel 4). Taken together, these observations strongly support the conclusion that the editing activity in the purified fractions from HeLa nuclear extracts is due entirely to the presence of hRED1 and that the human and rat RED1 proteins have a different substrate selectivity compared with dsRAD.
Figure 3.
Editing kinetics of the ds adenosine deaminases. (A) GluR-B Q/R site RNA editing by recombinant hRED1. The kinetics of the deaminase reactions were studied under single turnover reaction conditions ([protein] ≫ [GluR-B RNA]). The reaction was sampled at 0, 1, 3, 5, 10, 20, 30, 45, 60, and 90 min (lanes 1–10, respectively). For comparison, a parallel experiment with recombinant rRED1 is also shown. (B) Substrate selectivity of the ds adenosine deaminases. GluR-B RNA editing and dsRNA deamination activities were compared under single turnover reaction conditions. Increasing amounts of purified HeLa cell GluR-B activity (panel 1), recombinant hRED1 protein (panel 2), recombinant rRED1 protein (panel 3) and recombinant dsRAD protein (panel 4) were added to the reaction. The percentage of edited RNA was determined by thin layer chromatography and quantitated by the Fuji imaging system.
Substrate Requirements for RED1 Proteins.
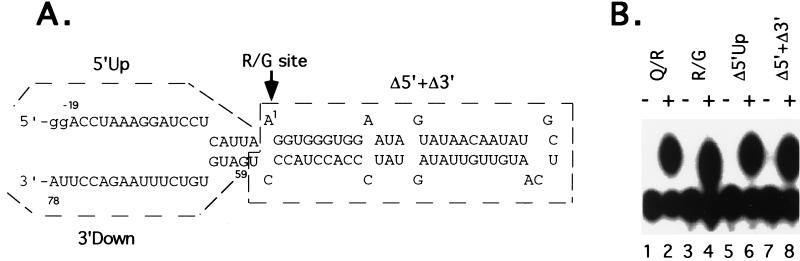
The RNA sequence and secondary structure of the GluR-B R/G site differs from the Q/R site. To investigate whether RED1 protein specifically edits the GluR-B R/G site, we determined the substrate requirements for RNA editing at the R/G site by recombinant rRED1. A 99-nucleotide RNA containing the R/G site, with 21 nucleotides upstream of the edited nucleotide and 78 nucleotides downstream (Fig. 4A), is edited efficiently by recombinant rRED1 (Fig. 4B, lanes 3 and 4). Previously, we demonstrated that RNA sequences immediately upstream from the edited adenosine residue at the Q/R site of GluR-B RNA are not required for editing (4). Interestingly, like the Q/R site, the nucleotides immediately upstream from the adenosine residue at the R/G site are not required for editing by the RED1 protein (Fig. 4B, Δ5′Up, lanes 5 and 6). Similarly, the sequence downstream of the ECS is not required for R/G site in vitro editing by RED1 (Fig. 4B, Δ5′Up + Δ3′Down, lanes 7 and 8). Thus an RNA of only 58 nucleotides, containing the editing site and an imperfect RNA stem structure, is sufficient for accurate R/G site editing.
Figure 4.
Editing of the minimal R/G substrate for rRED1. (A) RNA sequence and predicted structure surrounding the GluR-B R/G site. The minimal R/G site RNA consists of 19 nucleotides upstream of the R/G site (5′Up), 58 nucleotides of R/G site plus ECS sequence (Δ5′ + Δ3′) and 20 nucleotides downstream of the ECS sequence (3′Down). A dinucleotide (gg) was added to its 5′ end for efficient transcription. Arrow indicates the adenosine of the R/G editing site. (B) Editing activity was determined in the absence (−) or presence (+) of rRED1 for different lengths of R/G substrates. Q/R site RNA is used as a control (Q/R, lanes 1, 2). The minimal R/G site RNA (R/G, lanes 3 and 4) was made by RNA ligation of unlabeled 21 mer RNA (pppGGACCUAAAGGAUCCUCAUUA) and 5′ end-labeled 78 mer RNA lacking the 5′Up sequence (Δ5′Up). Deletion of 5′Up (Δ5′Up, lanes 5 and 6) and deletion of 5′Up and 3′Down (Δ5′ + Δ3′, lanes 7 and 8) were made by in vitro transcription based on PCR-generated templates. The adenosine at the editing site was added by ApG dinucleotide during transcription and labeled using T4 polynucleotide kinase.
To begin to understand the extent of the specificity of RED1 for the R/G site, we examined the effects of selected single base substitutions in areas of imperfect RNA duplex formation on in vitro editing with the rRED1 protein. Within the area of apparent RNA duplex, the minimal R/G site RNA contains an A–C mismatch at position 11 (A11C47) and G–G mismatch at position 15 (G15G43). Interestingly, the Q/R site RNA can form a similar structure near the editing site with mismatches at positions 13 (A13C309) and 20 (G20A302) (Fig. 5). Substitution of A11 of the R/G editing substrate or A13 of the Q/R substrate by a G had no effect on the in vitro editing activity, even though these mutations should stabilize the RNA stem structure by forming an additional base pair interaction. By contrast, when C47 of the R/G site or C309 of the Q/R site was changed to U, the efficiency of editing was reduced 2- to 3-fold. These mutations should also increase the stability of the RNA stem; however, they decrease the efficiency of editing, implying that base pairing at this position is not required for efficient editing. Rather, the editing activity displays a preference for specific sequence elements, preferring C to U at position 47 independent of base pairing. Double mutations in the R/G site RNA (C47 to U and G43 to C) that should result in a perfect RNA stem structure dramatically reduced the editing efficiency (40-fold). Similarly, double mutations at the corresponding sites in the Q/R site RNA (C309 to U and A302 to C) also reduced the efficiency of editing, in this case by ≈20-fold (Fig. 5A). Although the Q/R and R/G sites share a number of structural similarities, a difference occurs at the edited nucleotide. In the Q/R site this nucleotide is base-paired with its complement in the ECS, while at the R/G site there is a mismatch. This base pairing is not important for efficient Q/R editing in vivo (19). To investigate the role of this mismatch at the minimal R/G editing site, we synthesized an RNA with a C to U base substitution at position 57 in the R/G substrate. This substitution creates a base pair at the editing site, but had no significant effect on editing efficiency by rRED1 (A1U57 in Fig. 5B). Thus, for the end-labeled GluR-B substrate, base pairing at the R/G or Q/R editing sites is not required for efficient editing in vitro.
Figure 5.
The effect of mutations in the GluR-B Q/R and R/G site RNA on editing activity. (A) Editing activity of Q/R site mutations. (B) Editing activity of R/G site mutations. The potential secondary structure of the 58 nucleotide R/G site substrate and a similar 323 nucleotide Q/R site substrate are shown on the top of A and B. Two base mismatches at A13 and G20 of Q/R site, or A11 and G15 of the R/G site RNA were mutated to evaluate their importance for editing efficiency by rRED1. Editing efficiency was determined as in Fig. 3B. Mutations are denoted by the mismatched nucleotide and its position followed by its potential base paired nucleotide and its position. A13C309 and A11C47 are the wild-type substrates for Q/R and R/Q sites. In G13C309, nucleotide 13 of the Q/R site RNA is replaced by G. In A13U309, nucleotide 309 of Q/R site RNA is replaced by U. A13U309/G20C302 is a double mutation in which nucleotides 309 and 302 of the Q/R site RNA are replaced by U and C, respectively. The same nomenclature applies to the R/G site RNA mutations.
DISCUSSION
The GluR-B subunit of the mammalian glutamate receptor undergoes RNA editing at two specific adenosines that results in amino acid substitutions that profoundly alter the conductance properties of the ion channel (1, 12–13). A mutational analysis of the sequence determinants essential for efficient editing at the Q/R site demonstrated the requirement for RNA base pairing surrounding the editing site (4, 10). These data suggested that dsRAD might be responsible for the deamination of two specific adenosines in GluR-B pre-mRNAs (19). Our data, taken together with work from other labs (17–19), indicates that neither purified nor recombinant dsRAD is sufficient for accurate and efficient GluR-B editing at the Q/R site in vitro. However, these data could not exclude the possibility that dsRAD is responsible for GluR-B editing in vivo, but requires the presence of one or more cofactors that enhance the efficiency and specificity of editing (4, 17).
More recently, a second adenosine deaminase activity, RED1, was identified that shares significant homology with dsRAD. Recombinant RED1 can accurately and efficiently edit the Q/R site of GluR-B pre-mRNA in vitro (16). In this study, we provide independent evidence that GluR-B editing activity in vivo is mediated by RED1. We show that two distinct dsRNA-dependent adenosine deaminase activities in HeLa cell nuclear extracts can be separated by chromatography. One fraction reveals a nonspecific dsRNA adenosine deaminase activity, lacks specific GluR-B editing activity and contains dsRAD. The other HeLa cell activity specifically edits GluR-B Q/R RNA and contains RED1. We have cloned the hRED1 gene and expressed the hRED1 protein in a baculovirus system. The recombinant protein exhibits specific GluR-B editing at the Q/R and R/G sites and mimics the activity observed in extracts, suggesting that the RED1 protein alone can mediate efficient and specific GluR-B editing. Although the minimal R/G substrate (Fig. 4A) is not edited by dsRAD (data not shown), both RED1 and dsRAD can edit full length R/G site substrates with comparable efficiencies (16). Thus, we cannot rule out the possibility that the R/G site in GluR-B is edited by distinct deaminases in vivo.
We have also determined the substrate requirements for editing at the R/G site and compared these with the determinants of editing at the Q/R site. In initial experiments, we demonstrated that an RNA of only 58 nucleotides containing the R/G editing site and an imperfect RNA stem structure are sufficient for accurate R/G site editing. We found that editing prefers a C at position 47 for efficient editing, regardless of whether base pairing occurs at this position. Substitution of this C by a U in conjunction with a substitution of G43 with C creates a perfect duplex stem. This observation implies that dsRNA binding is not sufficient for site-specific editing. Rather, additional features such as specific sequences or mispaired nucleotides may be required. However, we do not know whether the loss of editing activity observed with the G43 mutation is a consequence of the loss of the mismatch at this position or the base substitution.
Finally, the widespread expression of both dsRAD and RED1 suggests that additional pre-mRNA substrates for these deaminase activities remain to be determined. HeLa cells, for example, express RED1 activity but do not express the glutamate receptor subunits. Identification of additional pre-mRNA substrates for these deaminase activities will provide additional insight into the mechanisms of RNA editing and the role of this process in regulating the differential expression of the protein isoforms that derive from edited and unedited mRNAs.
Acknowledgments
This work was supported by Grants GM42231 (to T.M.) and MH01079 (to P.S.) from the National Institutes of Health, and by funds from the Howard Hughes Medical Institute (to R.A). J.-H.Y was partially supported by the Chinese Natural Science Foundation.
ABBREVIATIONS
- GluR-B
glutamate receptor subunit B
- ECS
editing site complementary sequence
- ds
double stranded
References
- 1.Seeburg P H. Trends Neurosci. 1993;16:359–365. doi: 10.1016/0166-2236(93)90093-2. [DOI] [PubMed] [Google Scholar]
- 2.Cattano R. Curr Biol. 1994;4:134–136. doi: 10.1016/s0960-9822(94)00030-8. [DOI] [PubMed] [Google Scholar]
- 3.Chan L. BioEssays. 1993;15:33–41. doi: 10.1002/bies.950150106. [DOI] [PubMed] [Google Scholar]
- 4.Yang J-H, Sklar P, Axel R, Maniatis T. Nature (London) 1995;374:77–81. doi: 10.1038/374077a0. [DOI] [PubMed] [Google Scholar]
- 5.Melcher T, Maas S, Higuchi M, Keller W, Seeburg P H. J Biol Chem. 1995;270:8566–8570. doi: 10.1074/jbc.270.15.8566. [DOI] [PubMed] [Google Scholar]
- 6.Rueter S M, Burns C M, Coode S A, Mokherjee P, Emerson R B. Science. 1995;267:1491–1494. doi: 10.1126/science.7878468. [DOI] [PubMed] [Google Scholar]
- 7.Hume R I, Dingledine R, Heinemann S. Science. 1991;253:1028–1031. doi: 10.1126/science.1653450. [DOI] [PubMed] [Google Scholar]
- 8.Sommer B, Kohler M, Sprengel R, Seeburg P H. Cell. 1991;67:11–19. doi: 10.1016/0092-8674(91)90568-j. [DOI] [PubMed] [Google Scholar]
- 9.Burnashev N, Monyer H, Seeburg P H, Sakmann B. Neuron. 1992;8:189–198. doi: 10.1016/0896-6273(92)90120-3. [DOI] [PubMed] [Google Scholar]
- 10.Higuchi M, Single F N, Kohler M, Sommer B, Sprengel R, Seeburg P H. Cell. 1993;75:1361–1370. doi: 10.1016/0092-8674(93)90622-w. [DOI] [PubMed] [Google Scholar]
- 11.Herb A, Higuchi M, Sprengel R, Seeburg P H. Proc Natl Acad Sci USA. 1996;93:1875–1880. doi: 10.1073/pnas.93.5.1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hollmann M, Hartley M, Heinemann S. Science. 1991;252:851–853. doi: 10.1126/science.1709304. [DOI] [PubMed] [Google Scholar]
- 13.Verdoorn T A, Burnashev N, Monyer H, Seeburg P H, Sakmann B. Science. 1991;252:1715–1718. doi: 10.1126/science.1710829. [DOI] [PubMed] [Google Scholar]
- 14.Lomeli H, Mosbacher J, Melcher T, Hoger T, Geiger J R, Kuner T, Monyer H, Higuchi M, Bach A, Seeburg P H. Science. 1994;266:1709–1713. doi: 10.1126/science.7992055. [DOI] [PubMed] [Google Scholar]
- 15.Kohler M, Burnashev N, Sakmann B, Seeburg P H. Neuron. 1993;10:491–500. doi: 10.1016/0896-6273(93)90336-p. [DOI] [PubMed] [Google Scholar]
- 16.Melcher T, Maas S, Herb A, Sprengel R, Seeburg P H, Higuchi M. Nature (London) 1996;379:460–463. doi: 10.1038/379460a0. [DOI] [PubMed] [Google Scholar]
- 17.Dabiri G A, Lai F, Drakas R A, Nishikura K. EMBO J. 1996;15:34–45. [PMC free article] [PubMed] [Google Scholar]
- 18.Hurst S R, Hough R F, Aruscavage P J, Bass R L. RNA. 1995;1:1051–1060. [PMC free article] [PubMed] [Google Scholar]
- 19.Maas S, Melcher T, Herb A, Seeburg P H, Keller W, Krause S, Higuchi M, O’Connell M A. J Biol Chem. 1996;271:12221–12226. doi: 10.1074/jbc.271.21.12221. [DOI] [PubMed] [Google Scholar]