Abstract
Glucose tightly regulates the synthesis and secretion of insulin by β cells in the pancreatic islets of Langerhans. To investigate whether glucose regulates insulin synthesis at the level of insulin RNA splicing, we developed a method to detect and quantify a small amount of RNA by using the branched DNA (bDNA) signal-amplification technique. This assay is both sensitive and highly specific: mouse insulin II mRNA can be detected from a single β cell (βTC3 cells or mouse islets), whereas 1 million non-insulin-producing α cells (αTC1.6 cells) give no signal. By using intron and exon sequences, oligonucleotide probes were designed to distinguish the various unspliced and partially spliced insulin preRNAs from mature insulin mRNA. Insulin RNA splicing rates were estimated from the rate of disappearance of insulin preRNA signal from β cells treated with actinomycin D to block transcription. We found that the two introns in mouse insulin II are not spliced with the same efficiency. Intron 2 is spliced out more efficiently than intron 1. As a result, some mRNA retaining intron 1 enters the cytoplasm, making up ≈2-10% of insulin mRNA in the cell. This partially spliced cytoplasmic mRNA is quite stable, with a half-life similar to the completely spliced form. When islets grown in high glucose are shifted to low glucose medium, the level of insulin preRNA and the rate of splicing fall significantly. We conclude that glucose stimulates insulin gene transcription and insulin preRNA splicing. Previous estimates of insulin transcription rates based on insulin preRNA levels that did not consider the rate of splicing may have underestimated the effect of glucose on insulin gene transcription.
Insulin, the key signal for energy storage, is synthesized and secreted by the β cells of the pancreatic islets of Langerhans in mammals. Insulin synthesis and secretion is tightly regulated by glucose, which serves as a signal of the energy state of the organism. Synthesis of mature insulin is a multistep process and glucose regulates synthesis at several of these steps, including transcription (1), mRNA stabilization (2), translation (3, 4), and processing proinsulin to insulin (5).
The level of insulin mRNA available in the cytoplasm for translation to preproinsulin depends on the relative rates of insulin mRNA production and degradation. Glucose regulates both insulin gene transcription and insulin mRNA degradation (1, 2). Transcription rate alone, however, does not determine the production rate of cytoplasmic insulin mRNA. Prior to export from the nucleus, preRNAs are spliced and a 5′ m7GpppN cap and 3′ adenosines are added. These processes are not independent: splicing accelerates polyadenylylation, and both are important for nuclear export (6–8).
Insulin preRNA in most species contains two introns. The sequences of the introns show little evidence of evolutionary conservation, but the positions of the introns are highly conserved (9, 10). The length of intron 2 varies widely among species, but intron 1 is generally small (118 bp in mouse insulin II to 179 bp in human). Most species have a single insulin gene; but in rats and mice, a duplication has resulted in a second copy of the insulin gene that lacks the second intron (11–13). The ancestral and duplicate genes are termed insulin II and insulin I, respectively.
In the present study, we investigated the role of glucose in insulin RNA processing by measuring preRNA dynamics. Because preRNA levels are low and the insulin-producing β cells are not abundant, we developed a sensitive method for accurately measuring insulin RNAs. We adapted the branched DNA (bDNA) assay (Fig. 1) (14–16), a signal amplification technique, to measure the levels of insulin mRNA, preRNA, and preRNA splice products. We used this system to show that glucose regulates insulin preRNA splicing.
Figure 1.
Outline of bDNA assay. Short label and capture target probes are designed for a specific target sequence and hybridized with the target on a plate coated with oligonucleotides complementary to the capture target probes. The bDNA amplifier is then added and hybridized, followed by the AP probe, a short alkaline phosphatase-labeled oligonucleotide complementary to a triplicate repeat sequence on the bDNA branches. In the final step, a luminescent substrate of alkaline phosphatase, dioxetane, is added, and the emitted light is measured in a luminometer.
METHODS
Plasmid Construction and in Vitro RNA Transcription.
The mouse insulin I and mouse insulin II genomic DNA clones were obtained by PCR of mouse genomic DNA with primers complimentary to the exact ends of the primary transcripts of the two genes (13). The mouse insulin II cDNA clone was purified from βTC3 cDNA. PCR products were inserted into pBlueScript KS+. Mouse insulin I preRNA, mouse insulin II preRNA, and mouse insulin II mRNA was prepared from the linearized vectors using MEGAscript in vitro transcription kit for large-scale synthesis of RNA (Ambion, Austin, TX), by following the manufacturers instructions. RNA concentration was determined by measuring absorbance at 260 nm.
Cell Culture.
βTC3 and αTC 1.6 cells were cultured in Dulbecco’s modified Eagle’s medium with d-glucose (4.5 mg/liter), 15% horse serum, and 2.5% fetal bovine serum. For RNA decay experiments, actinomycin D1 was added at a concentration of 50 μg/ml to 106 βTC3 cells in 1 ml of medium on 35-mm culture dishes. At the time points shown, the cells were washed and lysed directly on the dishes in bDNA extraction buffer.
Mouse islet were picked by hand from collagenase-digested adult mouse pancreas (17), cultured in RPMI 1640 medium with 25 mM d-glucose and 10% fetal bovine serum for 4 days (18), and switched to either 25 mM or 2.5 mM glucose overnight prior to harvesting. RNA decay experiments were performed as for the βTC cells, except that cells were lysed in 1.5-ml microcentrifuge tubes.
Nuclear and cytoplasmic RNA was purified from 107 βTC3 cells by using the method of Dignam et al. (19) for purifying nuclear and cytoplasmic proteins, with the simple modification that protease inhibitors were replaced with RNase inhibitor. The entire procedure was performed rapidly on ice, and the cytoplasmic supernatant and pelleted nuclei were added directly to the bDNA assay.
Insulin bDNA Assay.
Insulin preRNA and mRNA were quantified using bDNA technology in a 96-microwell format similar to that described for quantification of hepatitis C virus RNA (16). All components, including buffers and DNA reagents were obtained from Chiron. Cells were lysed, or RNA samples were mixed, with 200 μl of extraction buffer [78 mM Hepes, pH 8.0/12.5 mM EDTA, pH 8.0/6.27 mM LiCl/1.6% lithium lauryl sulfate/proteinase K (1 mg/ml)/single-stranded DNA (19 μg/ml)/7.8% formamide/0.05% sodium azide/0.05% Proclin 300], along with proteinase K and 40 fmol of insulin-specific capture and label probes, loaded in the microwell plate, sealed with an adhesive-backed mylar Plate Sealer (Microtiter Plate Sealer; Flow Laboratory), and incubated overnight at 63°C in a plate heater to capture the targeted nucleic acids to the oligonucleotide-modified microwell surface. After cooling at room temperature for 10 min, wells were washed twice with wash A [0.1× standard saline citrate (SSC) and 0.1% SDS]. Fifty microliters of bDNA amplifier solution containing the bDNA amplifier at 1 pmol/ml in amplifier diluent [amplifier diluent was prepared by incubating 50% horse serum/1.3% SDS/6 mM Tris⋅HCl, pH 8.0/5× SSC/proteinase K (0.5 mg/ml) at 65°C for 2 hr, followed by addition of 1 mM phenylmethylsulfonyl fluoride to inactive the proteinase K and 0.05% sodium azide and 0.05% Proclin 300] was added and hybridized at 53°C for 30 min. After cooling and washing as described above, 50 μl of a mixture containing alkaline phosphatase-conjugated label probes (2 pmol/ml) in label diluent (amplifier diluent/0.1% Brij 35/1 mM ZnCl2/20 mM MgCl2) was added and hybridized at 53°C for 15 min. The plate was cooled and washed twice with wash A as above and then washed three times with wash B (0.1× SSC). Finally, 50 μl of chemiluminescent substrate (Lumiphos 530), an enzyme-triggerable dioxetane substrate for alkaline phosphatase, was added and the plate was incubated at 37°C for 25 min. Light emission was measured in a luminometer at 37°C.
The bDNA assay for insulin DNA was similarly performed in a 96-microwell format. The only difference from the RNA assay is in the first hybridization step, as described for quantification of hepatitis B virus DNA (20). Cells were lysed with 100 μl of extraction buffer in the microwell plate. The plate was incubated at 63°C in the plate heater for 30 min to digest and release the nucleic acids. fifty microliters of sodium hydroxide (final concentration, 0.17 M) mixed with 40 fmol of insulin-specific target probes were added and incubated at 63°C for another 30 min. After the DNA denaturation, 50 μl of neutralizing reagent (1 M 3-[N-morpholino]propanesulfonic acid/16× SSC/0.05% sodium azide) were added to the mixture. The samples were then incubated at 63°C overnight to capture the single-stranded insulin DNA molecules to the oligonucleotide-modified microwell surface. From this point the DNA assay is identical to the RNA assay.
Each sample was assayed in triplicate, and all data points represent the mean of at least three samples. RNA half-lives were calculated from the first three time points in Fig. 5B by assuming a first-order decay. At steady state, the transcription rate must equal the decay rate, which is calculated from the concentration and half-life of intron 2-containing preRNA.
Figure 5.
Glucose regulation of preRNA splicing. (A) The bDNA assay was performed with the probe sets shown and purified mouse islet cultured in 25 mM glucose for 4 days and then cultured in either 25 mM or 2.5 mM glucose overnight. The bars represent the ratio of the bDNA signal from islets grown at 2.5 or 25 mM glucose and assayed with the probe sets shown. (B) The bDNA assay was performed with the probe sets shown and purified mouse islet cultured in 25 mM glucose for 4 days and then cultured in either 25 mM or 2.5 mM glucose overnight. Islets were cultured in the presence of actinomycin D for the times indicated. Each data point represents the mean ± SEM of at least three samples.
The possibility that complex mixtures of RNA could interfere with the assay was tested by performing the assays of in vitro-transcribed synthetic RNA (Figs. 2A and 3B) with or without extracts from 106 αTC1.6 cells. The presence of the αTC1.6 cellular RNA did not affect the assay (data not shown).
Figure 2.
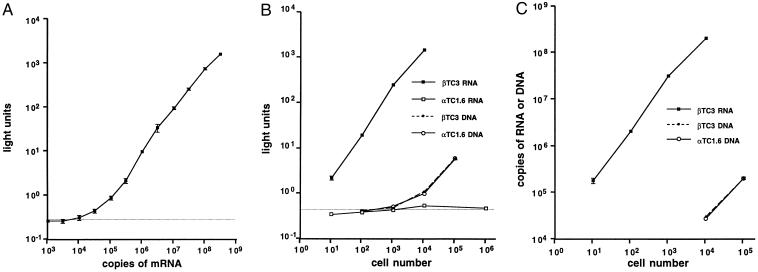
Sensitivity and specificity of the mouse insulin II mRNA assay. (A) The bDNA assay was performed with probe set A (see Fig. 3A) and in vitro-transcribed mouse insulin II mRNA. Data points represent the mean ± SD of at least three independent samples. (B) The bDNA assay for RNA or DNA was performed with probe set A and the cell types indicated. Data points represent the mean ± SEM of at least three samples. (C) Data in B were converted from light units to copies of RNA with the standard curve in A or copies of DNA with a similar standard curve for DNA. The dashed horizontal line in A and B represents the background light emission in the absence of RNA.
Figure 3.
Mouse insulin II preRNA assay. (A) The design of the preRNA probe sets is shown. The preRNA, probe positions, and probe number are accurately depicted to scale. Probes shown below the target sequence are capture target probes, and probes shown above the target are label target probes. (B) The bDNA assay was performed with the probe sets and the in vitro-transcribed RNAs shown. Data points represent the mean ± SEM of at least three samples. Bars below the dashed line are not significantly above the assay background.
RESULTS
The design of the bDNA assay is outlined in Fig. 1. Two sets of DNA oligonucleotides are synthesized. One set (capture probes) hybridizes to both the target sequence and to oligonucleotides linked to the assay plate. The second set (label probes) hybridizes to both the target sequence and the bDNA oligonucleotides (bDNA amplifiers). The final set of oligonucleotides (AP probe) is labeled with alkaline phosphatase and hybridizes to the branches of the amplifiers. When a luminescent substrate for alkaline phosphatase, dioxetane, is added, light is emitted. The net result is a marked signal amplification that is proportional to the concentration of the target sequence.
Different hybridization conditions allow DNA and RNA target sequences to be distinguished. If the sample is initially disrupted in an alkaline buffer, double-stranded DNA is denatured and RNA is destroyed. If the sample is harvested, instead, in a neutral buffer with SDS and proteinase K, DNA remains double-stranded and cannot hybridize with the probes and the RNA is protected from degradation.
Fig. 3A shows the design of the capture and label probes for mouse insulin II mRNA (oligonucleotide probe set A). Three capture probes target the 3′ end and 10 label probes target the remainder of the mRNA sequence.
RNA transcribed in vitro from mouse insulin II cDNA was used to determine the sensitivity of oligonucleotide set A (Fig. 2A). Light output increases linearly over four orders of magnitude of sample input. The limit of detection is approximately 104 insulin mRNA molecules. The optimal limit of detection for the bDNA assay is approximately 103 molecules (14), but the insulin mRNA assay is limited by a suboptimal number of capture and label probes because of the small size of the mouse insulin II mRNA.
In Fig. 2B, the signals are compared from two mouse islet tumor cell lines: the insulin-producing βTC3 cells (21) and the glucagon-producing αTC1.6 cells (22). For the βTC3 cells, the light signal in the RNA assay increases linearly with the number of cells used, with the limit of detection at approximately one cell. No signal is detected, however, by the RNA assay with αTCl.6 cells even when 106 cells are used. Measurements by the bDNA assay of GAPDH mRNA in the αTC1.6 and βTC3 cell extracts demonstrated that this difference was not due to difference in RNA recovery (data not shown). The DNA assay demonstrates that both cell types contain the insulin gene.
The standard curve in Fig. 2A was used to convert the RNA data in Fig. 2B to number of molecules (Fig. 2C). A similar standard curve for mouse insulin II genomic DNA was used to convert the DNA data. This data can be used to estimate the number of insulin II RNA molecules per βTC3 cell at approximately 2 × 104, consistent with previous evidence that βTC3 cells maintain high insulin mRNA levels (21). The estimate of gene copy is 2 copies per cell, as expected for a diploid cell.
Since 106 αTC-1.6 cells give no RNA signal, there must be fewer than 1 insulin II RNA molecule per 100 αTC-1.6 cells. By using this figure and the estimate of 2 × 104 copies per βTC3 cell, the difference in insulin II gene expression between these two closely related islet cell lines is greater than six orders of magnitude.
Probe set A was designed to target only the exon sequences of mouse insulin II RNA and, therefore, can capture any insulin II RNA, spliced or unspliced. Five other sets of oligonucleotide probes, sets B–F, were designed to recognize specific unspliced and partially spliced mouse insulin II preRNAs, by capturing and labeling exons and introns separately (Fig. 3A).
The specificities of the probe sets were tested using in vitro transcribed mouse insulin II mRNA, mouse insulin II preRNA, and mouse insulin I preRNA (Fig. 3B). There is no cross-reactivity of the preRNA sets (B–F) with the mature insulin II mRNA (Fig. 3B), even when 1 μg of the mRNA is used, whereas the A probe set gives a strong signal with both insulin II mRNA and preRNA. Although it gives a 10-fold weaker signal, the A probe set also recognizes the mouse insulin I sequences, reflecting the conservation of exon sequences between the two mouse insulin genes. Of the preRNA probe sets, only sets E and F, the sets that capture intron 1 sequences, cross-hybridize with mouse insulin I preRNA, and this signal is almost four orders of magnitude lower than the signal from mouse insulin II preRNA.
It is interesting to note that, although all probe sets recognize the mouse insulin I preRNA, the absolute light signals from different probe sets vary over a 10-fold range. Although part of this difference arises from the number of capture and label probes in each set, this difference also appears to depend on the grouping and hybridization characteristics of the individual oligonucleotide probes. The A set, for example, gives a stronger signal when tightly grouped on insulin II mRNA than when separated on the insulin II preRNA template. Because of these differences, a standard curve such as the one in Fig. 2A for probe set A with mouse insulin II mRNA was produced for each of the preRNA probe sets with mouse insulin II preRNA (data not shown).
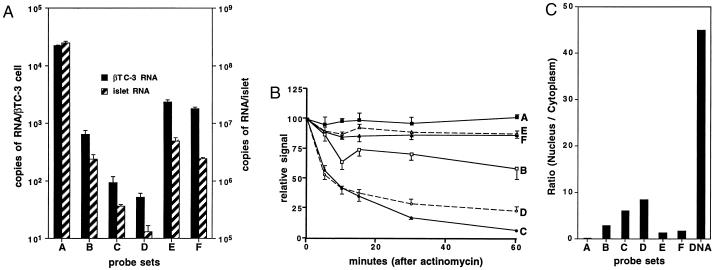
These standard curves were then used to calculate the abundance of different insulin II RNA splice forms in both βTC3 cells and cultured mouse islets (Fig. 4A). When an average of 2,000 β cells per mouse islet is assumed, these calculations estimate 105 insulin II mRNA per β cell. As expected, preRNA levels are lower; but the probe sets that recognize intron I containing RNAs (sets B, E, and F) give significantly higher estimates than the sets that recognize only intron 2 containing RNAs (sets C and D). Intron-1-containing RNA makes up 10% of the insulin II RNA in βTC3 cells and 2–3% in mouse islets. This high preRNA level did not appear to be due to cross-hybridization with noninsulin sequences, since none of the probe sets gave a signal with 106 αTC1.6 cells (data not shown).
Figure 4.
preRNA levels in β cells. (A) The bDNA assay was performed with the probe sets shown and either βTC3 cells (solid bars) or cultured mouse islets (hatched bars). Data points represent the mean ± SEM of at least three samples. (B) The bDNA assay was performed with the probe sets shown and βTC3 cells cultured in the presence of actinomycin D for the times indicated. Data points represent the mean ± SEM of at least three samples. (C) The bDNA assay was performed with the probe sets shown and cytoplasmic or nuclear extracts from βTC3 cells. Each bar labeled A through F represents the ratio of nuclear to cytoplasmic RNA levels; the bar labeled DNA depicts the ratio of nuclear to cytoplasmic DNA levels using the A probe.
Since insulin mRNA has a long half-life in β-cells (2), we assumed that the high level of intron-1-containing mRNA results from an extended half-life for this partially spliced RNA as well. This assumption was tested by treating the βTC-3 cells with actinomycin D to block transcription of new preRNA and measuring the fall in mRNA and preRNA levels (Fig. 4B). As expected, mature mRNA (probe set A) has a long half-life, well over 1 hr [greater than 48 hr by previous estimates (2)], and intron-2-containing RNA (probe sets D and C) has a very short half-life (approximately 7 min). Although there is an initial small drop in intron-1-containing RNA (probe sets B, E, and F), the half-lives of these partially spliced RNAs are close to that of mature mRNA.
Why does intron-1-containing RNA have such a long half-life? We hypothesized that partially spliced RNA containing intron 1 is exported from the nucleus to the cytoplasm where it can no longer be spliced and thus achieves a stability similar to fully spliced insulin II mRNA. This hypothesis is supported by the relatively low nuclear/cytoplasmic ratio of intron-1-containing RNA when compared with intron-2-containing RNA (Fig. 4C).
To test the ability of glucose to affect insulin II transcription and splicing rates, we cultured mouse islets at 25 mM or 2.5 mM glucose overnight for 16 hr, after several days of growth in 25 mM glucose. Insulin mRNA and intron-1-containing RNA levels decreased modestly after 16 hr at the lower glucose concentration whereas intron-2-containing RNA levels fell more sharply (Fig. 5A).
The rate of disappearance (splicing rate) of intron-2-containing preRNA increases with glucose concentration (Fig. 5B). The estimated half-life of intron-2-containing preRNA is 8.6 ± 0.5 min and 16.2 ± 1.7 min at 25 mM and 2.5 mM glucose, respectively (P value as calculated by paired Student’s t test <0.002). With this estimated change in splicing rate and the estimates of preRNA levels from Fig. 5A, it can be estimated that the rate of mouse insulin II gene transcription must fall approximately 17-fold when the glucose concentration decreases from 25 mM to 2.5 mM.
DISCUSSION
Our data demonstrate that the insulin bDNA assay is highly specific, precise, and sensitive to as few as 104 molecules of RNA. Although not as sensitive as the reverse transcriptase-coupled PCR assay, which can generally detect as few as 10 RNA molecules (23), the bDNA signal amplification method is more readily quantified than template amplification methods such as PCR. Also, because the target is not amplified as it is with PCR, cross-contamination with product sequences does not occur when performing multiple or repeated assays for the same target sequence.
Comparison with previous estimates of insulin RNA levels indicates that the assay is accurate as well. The estimate of 1 × 105 insulin II mRNA molecules per β cell in cultured islets is similar to the previous estimate of 50,000–150,000 insulin mRNAs per β cell by Northern blot hybridization with RNA from freshly isolated rat islets (24).
The finding that intron 1 of the insulin preRNA is inefficiently spliced, leading to the export of partially spliced insulin mRNA to the cytoplasm, was unexpected. Because intron 1 falls within the 5′ untranslated sequence of the insulin mRNA, its presence does not preclude the possibility that the mRNA is correctly translated to the preproinsulin peptide. Sequences within intron 1 have been shown to influence insulin gene transcription (25, 26), but the possible effects of intron 1 on translation have never been investigated. The presence of intron 1 could affect translation initiation, the overall rate of translation, or the regulation of translation by glucose and other regulators.
Why is intron 1 less efficiently spliced than intron 2? Two explanations seem possible: (i) Intron 1 is significantly smaller than intron 2, and smaller introns may be spliced less efficiently (27). (ii) The 5′ splice junction site for mouse insulin II intron 1 has only a 7- of 9-bp identity with the consensus 5′ splice sequence (28) for mammalian introns. The intron 2 5′ splice junction is a 9- of 9-bp match with the consensus sequence. When we sequenced multiple mouse insulin II cDNAs obtained from βTC3 cDNA by PCR, we found that the 5′ junction was variable, with the most common 5′ splice junction found 4 bp further downstream at a 6- of 9-bp match with the consensus sequence (data not shown). Aberrant intron 1 splicing of mouse insulin II RNA has been reported by others as well (29). Interestingly, the intron 1 splice donor sequence is poorly conserved among different species, and none of the known sequences form an ideal 5′ splice junction.
Inefficient splicing of intron 1 may explain the retention of intron 1 in the murine insulin I gene. The insulin I gene duplication is believed to have resulted from the integration of a cDNA produced by viral reverse transcription of an aberrant insulin II mRNA in a murine ancestor (30). We speculate that inefficient splicing of intron 1, such as we have observed, could explain why intron 1 was retained in the template mRNA.
Previous estimates of insulin gene transcription rate based on preRNA levels (31, 32) have underestimated the effect of glucose on transcription rate, because the splicing rate was not considered. The combination of accurate measurements of preRNA levels and preRNA removal (splicing) rates provided by the bDNA assay allows direct calculation of transcription rates, since at steady state, the rate of preRNA production (transcription) must equal the rate of removal. This calculation gives estimated transcription rates of 3.2 × 104 and 1.9 × 103 insulin II preRNA molecules per islet per min at 25 mM glucose and 2.5 mM glucose, respectively. This 17-fold difference in insulin gene transcription rates between high and low glucose is higher than the previous estimate of 6-fold calculated from nucleotide incorporation rates into insulin I mRNA in rat islets (1). The quantitative difference in the effect of glucose may derive from differences between the rat insulin I gene and the mouse insulin II gene, differences between rat and mouse islets, or the larger difference in glucose concentration used in our study [2.5 and 25 mM glucose vs. 3.3 and 17 mM glucose (1)].
Our results add preRNA splicing to the list of insulin synthetic processes that are regulated by glucose in β cells. These results do not distinguish, however, whether the effect of glucose on splicing is specific for the insulin preRNA or whether all β cell preRNAs are similarly regulated.
In summary, the use of the quantitative insulin bDNA assay has demonstrated that the two introns of the insulin gene are spliced with unequal efficiency and that insulin RNA splicing is regulated by glucose. This ability to accurately measure low levels of insulin mRNA and preRNA should allow for careful analysis of the physiologic regulation of insulin gene expression.
Acknowledgments
We thank I. F. German, Jr., for helpful discussions. This work was supported by National Institutes of Health Grant DK-41822 (M.S.G.) and a grant from the Diabetes Action Research and Education Foundation (M.S.G.).
ABBREVIATION
- bDNA
branched DNA
References
- 1.Nielsen D A, Welsh M, Casadaban M J, Steiner D F. J Biol Chem. 1985;260:13585–13589. [PubMed] [Google Scholar]
- 2.Welsh M, Nielsen D A, MacKrell A J, Steiner D F. J Biol Chem. 1985;260:13590–13594. [PubMed] [Google Scholar]
- 3.Welsh M, Scherberg N, Gilmore R, Steiner D F. Biochem J. 1986;235:459–467. doi: 10.1042/bj2350459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Permutt M A, Kipnis D M. J Biol Chem. 1972;247:1194–1199. [PubMed] [Google Scholar]
- 5.Nagamatsu S, Bolaffi J, Grodsky G. Endocrinology. 1987;120:1225–1231. doi: 10.1210/endo-120-4-1225. [DOI] [PubMed] [Google Scholar]
- 6.Legrain P, Rosbash M. Cell. 1989;57:573–583. doi: 10.1016/0092-8674(89)90127-x. [DOI] [PubMed] [Google Scholar]
- 7.Green M R. Curr Opin Cell Biol. 1989;1:519–525. doi: 10.1016/0955-0674(89)90014-8. [DOI] [PubMed] [Google Scholar]
- 8.Krug R M. Curr Opin Cell Biol. 1993;5:944–949. doi: 10.1016/0955-0674(93)90074-z. [DOI] [PubMed] [Google Scholar]
- 9.Steiner D F, Chan S J, Welsh J M, Kwok S C. Annu Rev Genet. 1985;19:463–484. doi: 10.1146/annurev.ge.19.120185.002335. [DOI] [PubMed] [Google Scholar]
- 10.Steiner D F, Chan S J. Horm Metab Res. 1988;20:443–444. doi: 10.1055/s-2007-1010855. [DOI] [PubMed] [Google Scholar]
- 11.Lomedico P, Rosenthal N, Efstratiadis A, Gilbert W, Kolodner R, Tizard R. Cell. 1979;18:545–558. doi: 10.1016/0092-8674(79)90071-0. [DOI] [PubMed] [Google Scholar]
- 12.Soares M B, Schon E, Henderson A, Karathanasis S K, Cate R, Zeitlin S, Chirgwin J, Efstratiadis A. Mol Cell Biol. 1985;5:2090–2103. doi: 10.1128/mcb.5.8.2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wentworth B M, Schaefer I M, Villa-Komaroff L, Chirgwin J M. J Mol Evol. 1986;23:305–312. doi: 10.1007/BF02100639. [DOI] [PubMed] [Google Scholar]
- 14.Urdea M S, Running J, Horn T, Clyne J, Ku L, Warner B. Gene. 1987;61:253–264. doi: 10.1016/0378-1119(87)90189-2. [DOI] [PubMed] [Google Scholar]
- 15.Urdea M S, Kolberg J, Warner B, Horn T, Clyne J, Ku L, Running J. In: Luminescence Immunoassay and Molecular Applications. Van Dyke R, editor. 1990. pp. 276–292. [Google Scholar]
- 16.Davis G L, Lau J Y, Urdea M S, Neuwald P D, Wilber J C, Lindsay K, Perrillo R P, Albrecht J. Hepatology. 1994;19:1337–1341. [PubMed] [Google Scholar]
- 17.Lacy P E, Kostianovsky M. Diabetes. 1967;16:35–39. doi: 10.2337/diab.16.1.35. [DOI] [PubMed] [Google Scholar]
- 18.Liang Y, Najafi H, Matschinsky F M. J Biol Chem. 1990;265:16863–6. [PubMed] [Google Scholar]
- 19.Dignam J D, Lebowitz R M, Roeder R G. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hendricks D A, Stowe B J, Hoo B S, Kolberg J, Irvine B D, Neuwald P D, Urdea M S, Perrillo R P. Am J Clin Pathol. 1995;104:537–546. doi: 10.1093/ajcp/104.5.537. [DOI] [PubMed] [Google Scholar]
- 21.Efrat S, Linde S, Kofod H, Spector D, Delannoy M, Grant S, Hanahan D, Baekkeskov S. Proc Natl Acad Sci USA. 1988;85:9037–9041. doi: 10.1073/pnas.85.23.9037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hamaguchi K, Leiter E. Diabetes. 1990;39:415–425. doi: 10.2337/diab.39.4.415. [DOI] [PubMed] [Google Scholar]
- 23.Rappolee D A, Mark D, Banda M J, Werb Z. Science. 1988;241:708–712. doi: 10.1126/science.3041594. [DOI] [PubMed] [Google Scholar]
- 24.Giddings S J, Chirgwin J, Permutt M A. Diabetologia. 1985;28:343–347. doi: 10.1007/BF00283141. [DOI] [PubMed] [Google Scholar]
- 25.Inagaki N, Maekawa T, Sudo T, Ishii S, Seino Y, Imura H. Proc Natl Acad Sci USA. 1992;89:1045–1049. doi: 10.1073/pnas.89.3.1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reibel L, Besnard C, Lores P, Jami J, Gacon G. Nucleic Acids Res. 1993;21:1595–1600. doi: 10.1093/nar/21.7.1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fu X Y, Manley J L. Mol Cell Biol. 1987;7:738–748. doi: 10.1128/mcb.7.2.738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mount S M. Nucleic Acids Res. 1982;10:459–472. doi: 10.1093/nar/10.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Neophytou P I, Muir E M, Hutton J C. Diabetes. 1996;45:127–33. doi: 10.2337/diab.45.2.127. [DOI] [PubMed] [Google Scholar]
- 30.Soares M, Schon E, Henderson A, Karathanasis S K, Cate R, Zeitlin S, Chirgwin J, Efstratiadis A. Mol Cell Biol. 1985;5:2099–2103. doi: 10.1128/mcb.5.8.2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Giddings S J, Carnaghi L R. J Biol Chem. 1988;263:3845–3849. [PubMed] [Google Scholar]
- 32.Giddings S J, Carnaghi L R, Fischer L J, Miller C P. Mol Endocrinol. 1991;5:549–554. doi: 10.1210/mend-5-4-549. [DOI] [PubMed] [Google Scholar]