Abstract
A combined physical and genetic map of the chromosome of Lactococcus lactis subsp. lactis IL1403 was determined. We constructed a restriction map for the NotI, ApaI, and SmaI enzymes. The order of the restriction fragments was determined by using the randomly integrative plasmid pRL1 and by performing indirect end-labeling experiments. The strain IL1403 chromosome was found to be circular and 2,420 kb in size. A total of 24 chromosomal markers were mapped on the chromosome by performing hybridization experiments with gene probes for L. lactis and various other bacteria. Integration of pRC1-derived plasmids via homologous recombination allowed more precise location of some lactococcal genes and allowed us to determine the orientation of these genes on the chromosome. Recurrent sequences, such as insertion elements and rRNA gene (rrn) clusters, were also mapped. At least seven copies of IS1076 were present and were located on 50% of the chromosome. In contrast, no copy of ISS1RS was detected. Six ribosomal operons were found on the strain IL1403 chromosome; five were located on 16% of the chromosome and were transcribed in the same direction. A comparison of the physical maps of L. lactis subsp. lactis IL1403 and DL11 showed that these two strains are closely related and that the variable regions are located mainly near the rrn gene clusters. In contrast, despite major restriction pattern dissimilarities between L. lactis IL1403 and MG1363, the overall genetic organization of the genome seems to be conserved between these two strains.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aguinaga M. del P., Sharan C., Singh D. N., Valenzuela M. S. Efficient transfer of DNA to nylon membranes using vacuum blotting in presence of alkali. Biotechniques. 1989 Nov-Dec;7(10):1077–1079. [PubMed] [Google Scholar]
- Albertsen H. M., Le Paslier D., Abderrahim H., Dausset J., Cann H., Cohen D. Improved control of partial DNA restriction enzyme digest in agarose using limiting concentrations of Mg++. Nucleic Acids Res. 1989 Jan 25;17(2):808–808. doi: 10.1093/nar/17.2.808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allardet-Servent A., Carles-Nurit M. J., Bourg G., Michaux S., Ramuz M. Physical map of the Brucella melitensis 16 M chromosome. J Bacteriol. 1991 Apr;173(7):2219–2224. doi: 10.1128/jb.173.7.2219-2224.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alloing G., Trombe M. C., Claverys J. P. The ami locus of the gram-positive bacterium Streptococcus pneumoniae is similar to binding protein-dependent transport operons of gram-negative bacteria. Mol Microbiol. 1990 Apr;4(4):633–644. doi: 10.1111/j.1365-2958.1990.tb00632.x. [DOI] [PubMed] [Google Scholar]
- Anderson P., Roth J. Spontaneous tandem genetic duplications in Salmonella typhimurium arise by unequal recombination between rRNA (rrn) cistrons. Proc Natl Acad Sci U S A. 1981 May;78(5):3113–3117. doi: 10.1073/pnas.78.5.3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmann B. J. Linkage map of Escherichia coli K-12, edition 8. Microbiol Rev. 1990 Jun;54(2):130–197. doi: 10.1128/mr.54.2.130-197.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bancroft I., Wolk C. P., Oren E. V. Physical and genetic maps of the genome of the heterocyst-forming cyanobacterium Anabaena sp. strain PCC 7120. J Bacteriol. 1989 Nov;171(11):5940–5948. doi: 10.1128/jb.171.11.5940-5948.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardowski J., Ehrlich S. D., Chopin A. Tryptophan biosynthesis genes in Lactococcus lactis subsp. lactis. J Bacteriol. 1992 Oct;174(20):6563–6570. doi: 10.1128/jb.174.20.6563-6570.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beresford T., Condon S. Cloning and partial characterization of genes for ribosomal ribonucleic acid in Lactococcus lactis subsp. lactis. FEMS Microbiol Lett. 1991 Mar 1;62(2-3):319–323. doi: 10.1016/0378-1097(91)90178-d. [DOI] [PubMed] [Google Scholar]
- Birkenbihl R. P., Vielmetter W. Complete maps of IS1, IS2, IS3, IS4, IS5, IS30 and IS150 locations in Escherichia coli K12. Mol Gen Genet. 1989 Dec;220(1):147–153. doi: 10.1007/BF00260869. [DOI] [PubMed] [Google Scholar]
- Butler P. D., Moxon E. R. A physical map of the genome of Haemophilus influenzae type b. J Gen Microbiol. 1990 Dec;136(12):2333–2342. doi: 10.1099/00221287-136-12-2333. [DOI] [PubMed] [Google Scholar]
- Canard B., Cole S. T. Genome organization of the anaerobic pathogen Clostridium perfringens. Proc Natl Acad Sci U S A. 1989 Sep;86(17):6676–6680. doi: 10.1073/pnas.86.17.6676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canard B., Saint-Joanis B., Cole S. T. Genomic diversity and organization of virulence genes in the pathogenic anaerobe Clostridium perfringens. Mol Microbiol. 1992 Jun;6(11):1421–1429. doi: 10.1111/j.1365-2958.1992.tb00862.x. [DOI] [PubMed] [Google Scholar]
- Chang N., Taylor D. E. Use of pulsed-field agarose gel electrophoresis to size genomes of Campylobacter species and to construct a SalI map of Campylobacter jejuni UA580. J Bacteriol. 1990 Sep;172(9):5211–5217. doi: 10.1128/jb.172.9.5211-5217.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chopin A., Chopin M. C., Moillo-Batt A., Langella P. Two plasmid-determined restriction and modification systems in Streptococcus lactis. Plasmid. 1984 May;11(3):260–263. doi: 10.1016/0147-619x(84)90033-7. [DOI] [PubMed] [Google Scholar]
- Chopin M. C., Chopin A., Rouault A., Galleron N. Insertion and amplification of foreign genes in the Lactococcus lactis subsp. lactis chromosome. Appl Environ Microbiol. 1989 Jul;55(7):1769–1774. doi: 10.1128/aem.55.7.1769-1774.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels D. L. The complete AvrII restriction map of the Escherichia coli genome and comparisons of several laboratory strains. Nucleic Acids Res. 1990 May 11;18(9):2649–2651. doi: 10.1093/nar/18.9.2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delorme C., Ehrlich S. D., Renault P. Histidine biosynthesis genes in Lactococcus lactis subsp. lactis. J Bacteriol. 1992 Oct;174(20):6571–6579. doi: 10.1128/jb.174.20.6571-6579.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garnier T., Canard B., Cole S. T. Cloning, mapping, and molecular characterization of the rRNA operons of Clostridium perfringens. J Bacteriol. 1991 Sep;173(17):5431–5438. doi: 10.1128/jb.173.17.5431-5438.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasc A. M., Kauc L., Barraillé P., Sicard M., Goodgal S. Gene localization, size, and physical map of the chromosome of Streptococcus pneumoniae. J Bacteriol. 1991 Nov;173(22):7361–7367. doi: 10.1128/jb.173.22.7361-7367.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasson M. J. Plasmid complements of Streptococcus lactis NCDO 712 and other lactic streptococci after protoplast-induced curing. J Bacteriol. 1983 Apr;154(1):1–9. doi: 10.1128/jb.154.1.1-9.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godon J. J., Chopin M. C., Ehrlich S. D. Branched-chain amino acid biosynthesis genes in Lactococcus lactis subsp. lactis. J Bacteriol. 1992 Oct;174(20):6580–6589. doi: 10.1128/jb.174.20.6580-6589.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grothues D., Tümmler B. New approaches in genome analysis by pulsed-field gel electrophoresis: application to the analysis of Pseudomonas species. Mol Microbiol. 1991 Nov;5(11):2763–2776. doi: 10.1111/j.1365-2958.1991.tb01985.x. [DOI] [PubMed] [Google Scholar]
- Huang D. C., Novel M., Novel G. A transposon-like element on the lactose plasmid of Lactococcus lactis subsp. lactis Z270. FEMS Microbiol Lett. 1991 Jan 1;61(1):101–106. doi: 10.1016/0378-1097(91)90021-2. [DOI] [PubMed] [Google Scholar]
- Jarvis A. W., Fitzgerald G. F., Mata M., Mercenier A., Neve H., Powell I. B., Ronda C., Saxelin M., Teuber M. Species and type phages of lactococcal bacteriophages. Intervirology. 1991;32(1):2–9. doi: 10.1159/000150179. [DOI] [PubMed] [Google Scholar]
- Kok J., Venema G. Genetics of proteinases of lactic acid bacteria. Biochimie. 1988 Apr;70(4):475–488. doi: 10.1016/0300-9084(88)90084-3. [DOI] [PubMed] [Google Scholar]
- Krawiec S., Riley M. Organization of the bacterial chromosome. Microbiol Rev. 1990 Dec;54(4):502–539. doi: 10.1128/mr.54.4.502-539.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler G., Ludwig W., Schleifer K. H. Differentiation of lactococci by rRNA gene restriction analysis. FEMS Microbiol Lett. 1991 Dec 1;68(3):307–312. doi: 10.1111/j.1574-6968.1991.tb04615.x. [DOI] [PubMed] [Google Scholar]
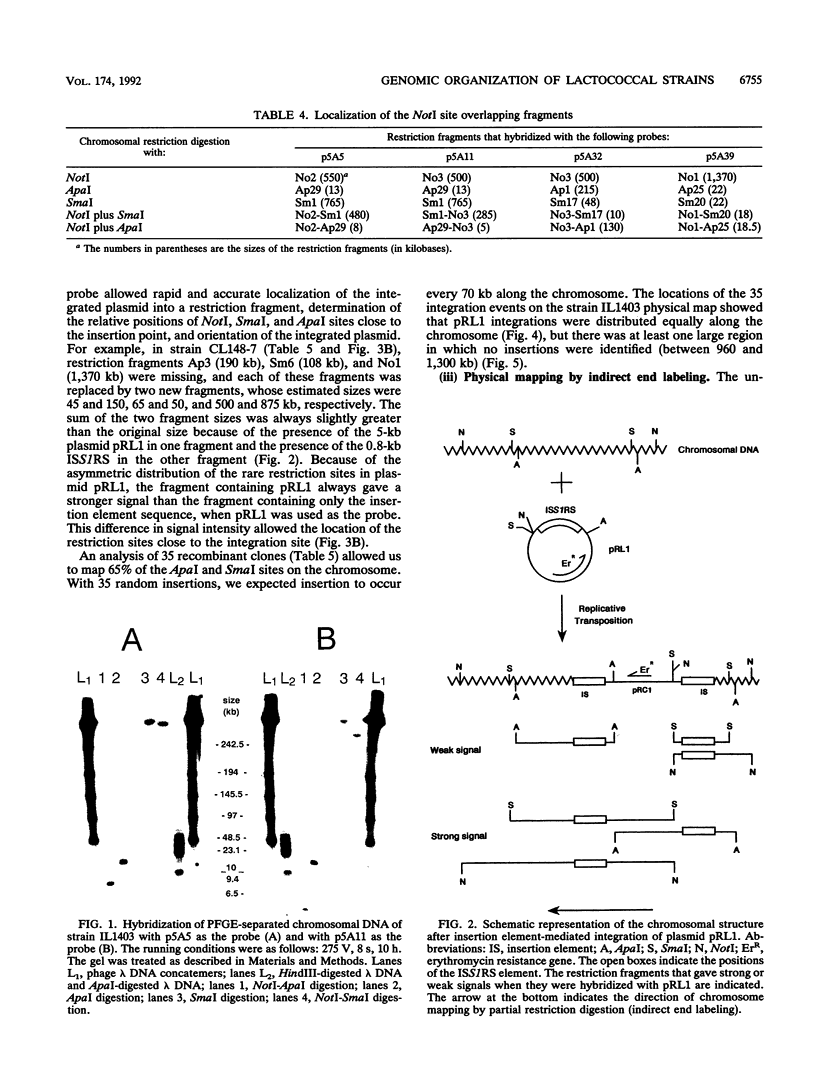
- Le Bourgeois P., Lautier M., Mata M., Ritzenthaler P. New tools for the physical and genetic mapping of Lactococcus strains. Gene. 1992 Feb 1;111(1):109–114. doi: 10.1016/0378-1119(92)90610-2. [DOI] [PubMed] [Google Scholar]
- Le Bourgeois P., Mata M., Ritzenthaler P. Genome comparison of Lactococcus strains by pulsed-field gel electrophoresis. FEMS Microbiol Lett. 1989 May;50(1-2):65–69. doi: 10.1016/0378-1097(89)90460-6. [DOI] [PubMed] [Google Scholar]
- LeBlanc D. J., Crow V. L., Lee L. N., Garon C. F. Influence of the lactose plasmid on the metabolism of galactose by Streptococcus lactis. J Bacteriol. 1979 Feb;137(2):878–884. doi: 10.1128/jb.137.2.878-884.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leenhouts K. J., Kok J., Venema G. Stability of Integrated Plasmids in the Chromosome of Lactococcus lactis. Appl Environ Microbiol. 1990 Sep;56(9):2726–2735. doi: 10.1128/aem.56.9.2726-2735.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauël C., Young M., Margot P., Karamata D. The essential nature of teichoic acids in Bacillus subtilis as revealed by insertional mutagenesis. Mol Gen Genet. 1989 Feb;215(3):388–394. doi: 10.1007/BF00427034. [DOI] [PubMed] [Google Scholar]
- Muto A., Osawa S. The guanine and cytosine content of genomic DNA and bacterial evolution. Proc Natl Acad Sci U S A. 1987 Jan;84(1):166–169. doi: 10.1073/pnas.84.1.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Méjean V., Devedjian J. C., Rives I., Alloing G., Claverys J. P. Uracil-DNA glycosylase affects mismatch repair efficiency in transformation and bisulfite-induced mutagenesis in Streptococcus pneumoniae. Nucleic Acids Res. 1991 Oct 25;19(20):5525–5531. doi: 10.1093/nar/19.20.5525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nardi M., Chopin M. C., Chopin A., Cals M. M., Gripon J. C. Cloning and DNA sequence analysis of an X-prolyl dipeptidyl aminopeptidase gene from Lactococcus lactis subsp. lactis NCDO 763. Appl Environ Microbiol. 1991 Jan;57(1):45–50. doi: 10.1128/aem.57.1.45-50.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novel M., Huang X. F., Novel G. Cloning of a chromosomal fragment from Lactococcus lactis subsp. lactis partially complementing Escherichia coli recA functions. FEMS Microbiol Lett. 1990 Nov;60(3):309–314. doi: 10.1016/0378-1097(90)90323-i. [DOI] [PubMed] [Google Scholar]
- Nuijten P. J., Bartels C., Bleumink-Pluym N. M., Gaastra W., van der Zeijst B. A. Size and physical map of the Campylobacter jejuni chromosome. Nucleic Acids Res. 1990 Nov 11;18(21):6211–6214. doi: 10.1093/nar/18.21.6211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okahashi N., Sasakawa C., Okada N., Yamada M., Yoshikawa M., Tokuda M., Takahashi I., Koga T. Construction of NotI restriction map of the Streptococcus mutans genome. J Gen Microbiol. 1990 Nov;136(11):2217–2223. doi: 10.1099/00221287-136-11-2217. [DOI] [PubMed] [Google Scholar]
- Piggot P. J., Hoch J. A. Revised genetic linkage map of Bacillus subtilis. Microbiol Rev. 1985 Jun;49(2):158–179. doi: 10.1128/mr.49.2.158-179.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polzin K. M., McKay L. L. Identification, DNA sequence, and distribution of IS981, a new, high-copy-number insertion sequence in lactococci. Appl Environ Microbiol. 1991 Mar;57(3):734–743. doi: 10.1128/aem.57.3.734-743.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell Ian B., Achen Marc G., Hillier Alan J., Davidson Barrie E. A Simple and Rapid Method for Genetic Transformation of Lactic Streptococci by Electroporation. Appl Environ Microbiol. 1988 Mar;54(3):655–660. doi: 10.1128/aem.54.3.655-660.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price C. W., Gitt M. A., Doi R. H. Isolation and physical mapping of the gene encoding the major sigma factor of Bacillus subtilis RNA polymerase. Proc Natl Acad Sci U S A. 1983 Jul;80(13):4074–4078. doi: 10.1073/pnas.80.13.4074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prudhomme M., Méjean V., Martin B., Claverys J. P. Mismatch repair genes of Streptococcus pneumoniae: HexA confers a mutator phenotype in Escherichia coli by negative complementation. J Bacteriol. 1991 Nov;173(22):7196–7203. doi: 10.1128/jb.173.22.7196-7203.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renault P., Gaillardin C., Heslot H. Product of the Lactococcus lactis gene required for malolactic fermentation is homologous to a family of positive regulators. J Bacteriol. 1989 Jun;171(6):3108–3114. doi: 10.1128/jb.171.6.3108-3114.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross P., O'Gara F., Condon S. Cloning and characterization of the thymidylate synthase gene from Lactococcus lactis subsp. lactis. Appl Environ Microbiol. 1990 Jul;56(7):2156–2163. doi: 10.1128/aem.56.7.2156-2163.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross P., O'Gara F., Condon S. Thymidylate synthase gene from Lactococcus lactis as a genetic marker: an alternative to antibiotic resistance genes. Appl Environ Microbiol. 1990 Jul;56(7):2164–2169. doi: 10.1128/aem.56.7.2164-2169.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz D. C., Cantor C. R. Separation of yeast chromosome-sized DNAs by pulsed field gradient gel electrophoresis. Cell. 1984 May;37(1):67–75. doi: 10.1016/0092-8674(84)90301-5. [DOI] [PubMed] [Google Scholar]
- Schäfer A., Jahns A., Geis A., Teuber M. Distribution of the IS elements ISS1 and IS904 in lactococci. FEMS Microbiol Lett. 1991 May 15;64(2-3):311–317. doi: 10.1111/j.1574-6968.1991.tb04681.x. [DOI] [PubMed] [Google Scholar]
- Sitzmann J., Klein A. Physical and genetic map of the Methanococcus voltae chromosome. Mol Microbiol. 1991 Feb;5(2):505–513. doi: 10.1111/j.1365-2958.1991.tb02134.x. [DOI] [PubMed] [Google Scholar]
- Stackebrandt E., Teuber M. Molecular taxonomy and phylogenetic position of lactic acid bacteria. Biochimie. 1988 Mar;70(3):317–324. doi: 10.1016/0300-9084(88)90204-0. [DOI] [PubMed] [Google Scholar]
- Steen M. T., Chung Y. J., Hansen J. N. Characterization of the nisin gene as part of a polycistronic operon in the chromosome of Lactococcus lactis ATCC 11454. Appl Environ Microbiol. 1991 Apr;57(4):1181–1188. doi: 10.1128/aem.57.4.1181-1188.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh J. W., Boylan S. A., Price C. W. Gene for the alpha subunit of Bacillus subtilis RNA polymerase maps in the ribosomal protein gene cluster. J Bacteriol. 1986 Oct;168(1):65–71. doi: 10.1128/jb.168.1.65-71.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanskanen E. I., Tulloch D. L., Hillier A. J., Davidson B. E. Pulsed-Field Gel Electrophoresis of SmaI Digests of Lactococcal Genomic DNA, a Novel Method of Strain Identification. Appl Environ Microbiol. 1990 Oct;56(10):3105–3111. doi: 10.1128/aem.56.10.3105-3111.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terzaghi B. E., Sandine W. E. Improved medium for lactic streptococci and their bacteriophages. Appl Microbiol. 1975 Jun;29(6):807–813. doi: 10.1128/am.29.6.807-813.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tulloch D. L., Finch L. R., Hillier A. J., Davidson B. E. Physical map of the chromosome of Lactococcus lactis subsp. lactis DL11 and localization of six putative rRNA operons. J Bacteriol. 1991 May;173(9):2768–2775. doi: 10.1128/jb.173.9.2768-2775.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umeda M., Ohtsubo E. Mapping of insertion elements IS1, IS2 and IS3 on the Escherichia coli K-12 chromosome. Role of the insertion elements in formation of Hfrs and F' factors and in rearrangement of bacterial chromosomes. J Mol Biol. 1989 Aug 20;208(4):601–614. doi: 10.1016/0022-2836(89)90151-4. [DOI] [PubMed] [Google Scholar]
- Vogelstein B., Gillespie D. Preparative and analytical purification of DNA from agarose. Proc Natl Acad Sci U S A. 1979 Feb;76(2):615–619. doi: 10.1073/pnas.76.2.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterbury P. G., Lane M. J. Generation of lambda phage concatemers for use as pulsed field electrophoresis size markers. Nucleic Acids Res. 1987 May 11;15(9):3930–3930. doi: 10.1093/nar/15.9.3930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitaker R. D., Batt C. A. Characterization of the Heat Shock Response in Lactococcus lactis subsp. lactis. Appl Environ Microbiol. 1991 May;57(5):1408–1412. doi: 10.1128/aem.57.5.1408-1412.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong K. K., McClelland M. A BlnI restriction map of the Salmonella typhimurium LT2 genome. J Bacteriol. 1992 Mar;174(5):1656–1661. doi: 10.1128/jb.174.5.1656-1661.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeigler D. R., Dean D. H. Orientation of genes in the Bacillus subtilis chromosome. Genetics. 1990 Aug;125(4):703–708. doi: 10.1093/genetics/125.4.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Asseldonk M., Rutten G., Oteman M., Siezen R. J., de Vos W. M., Simons G. Cloning of usp45, a gene encoding a secreted protein from Lactococcus lactis subsp. lactis MG1363. Gene. 1990 Oct 30;95(1):155–160. doi: 10.1016/0378-1119(90)90428-t. [DOI] [PubMed] [Google Scholar]