Abstract
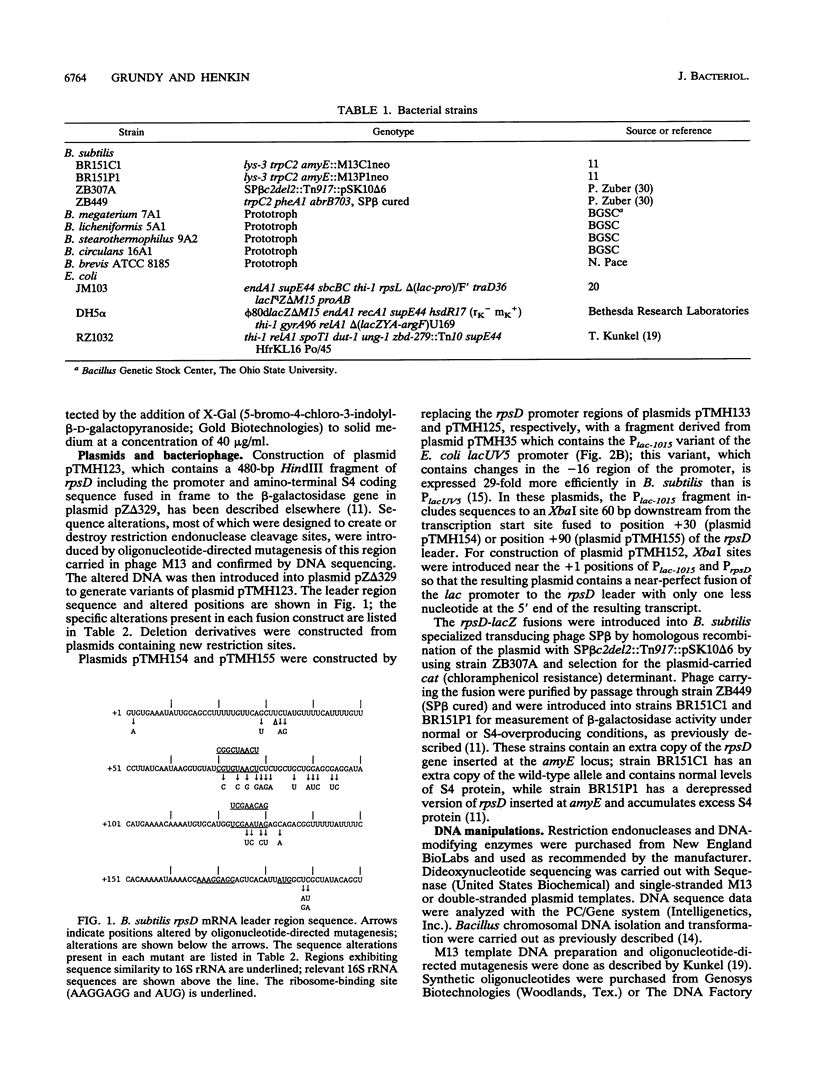
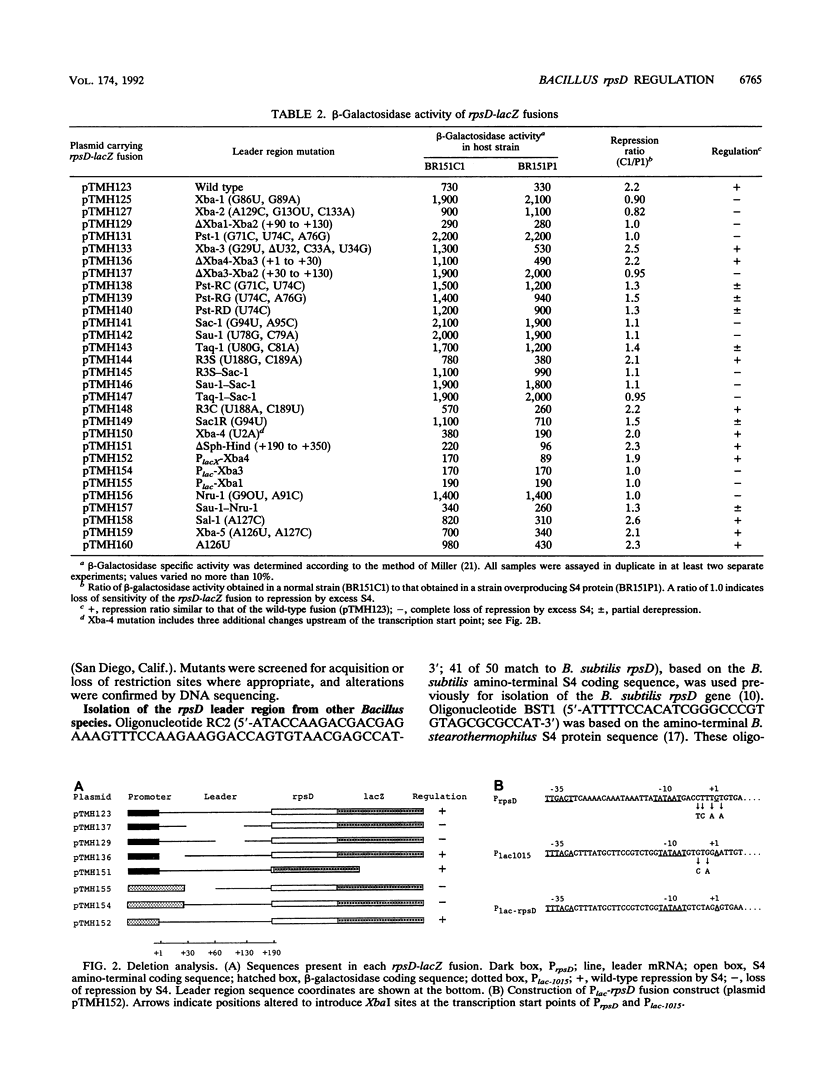
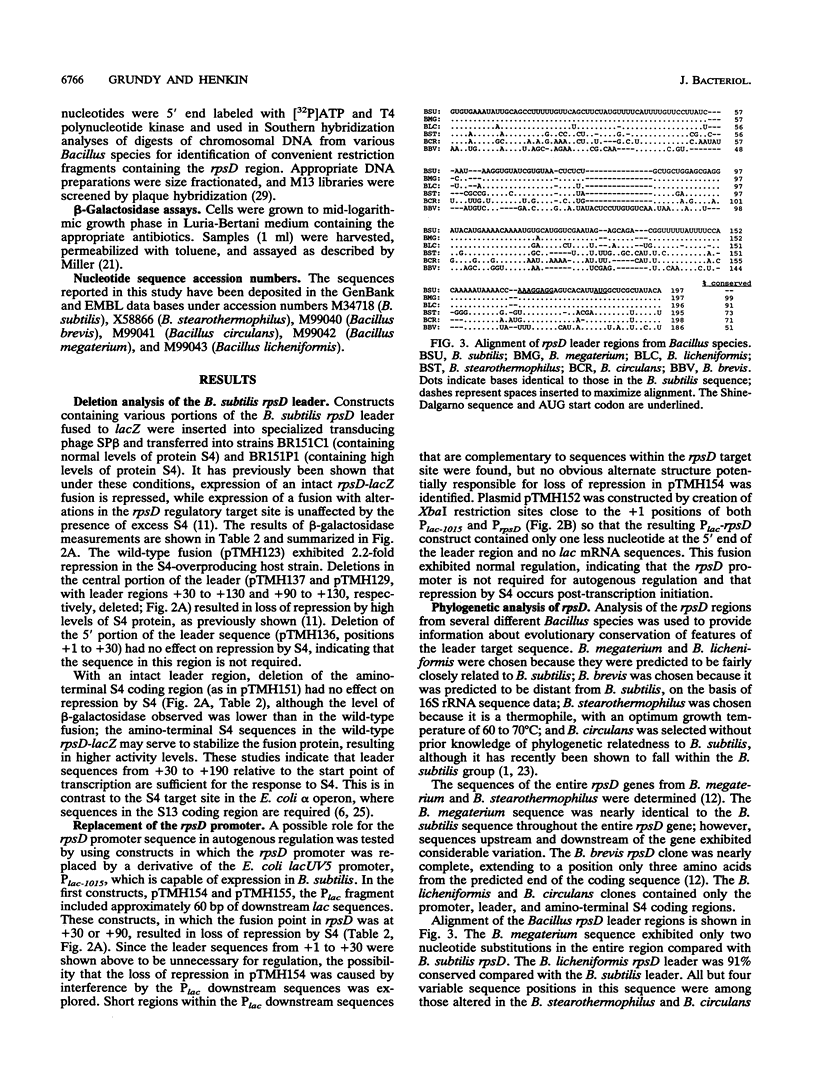
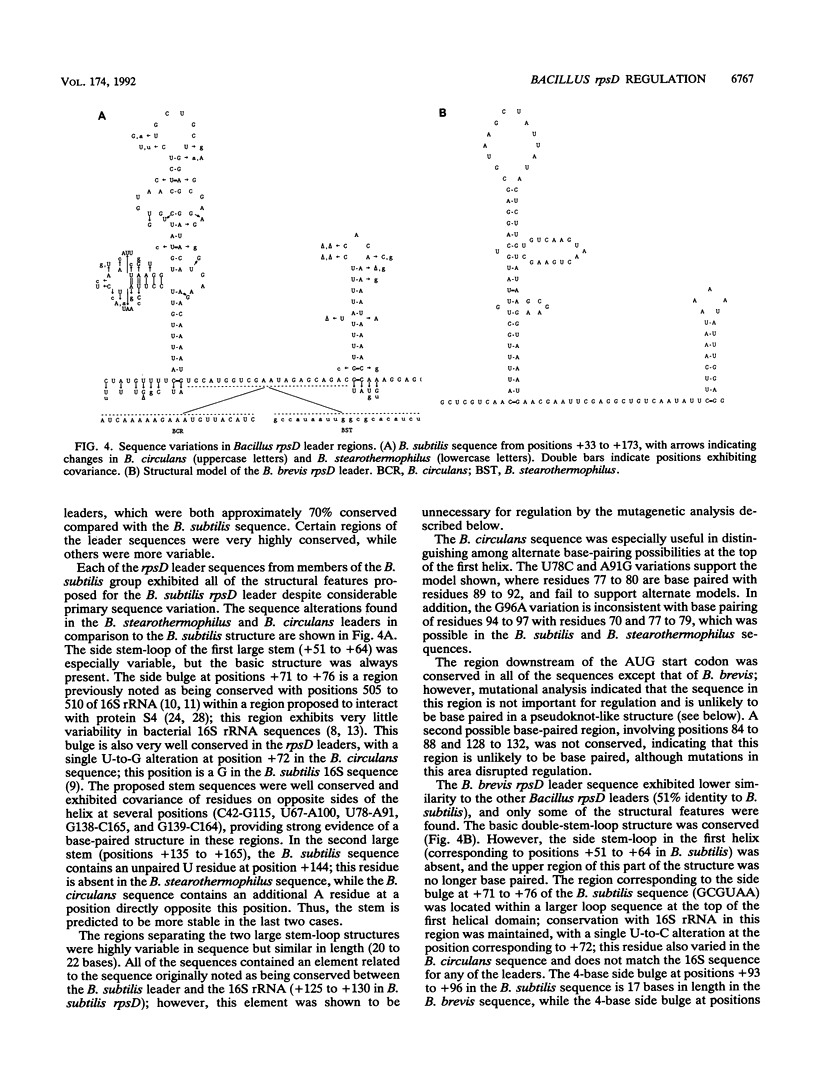
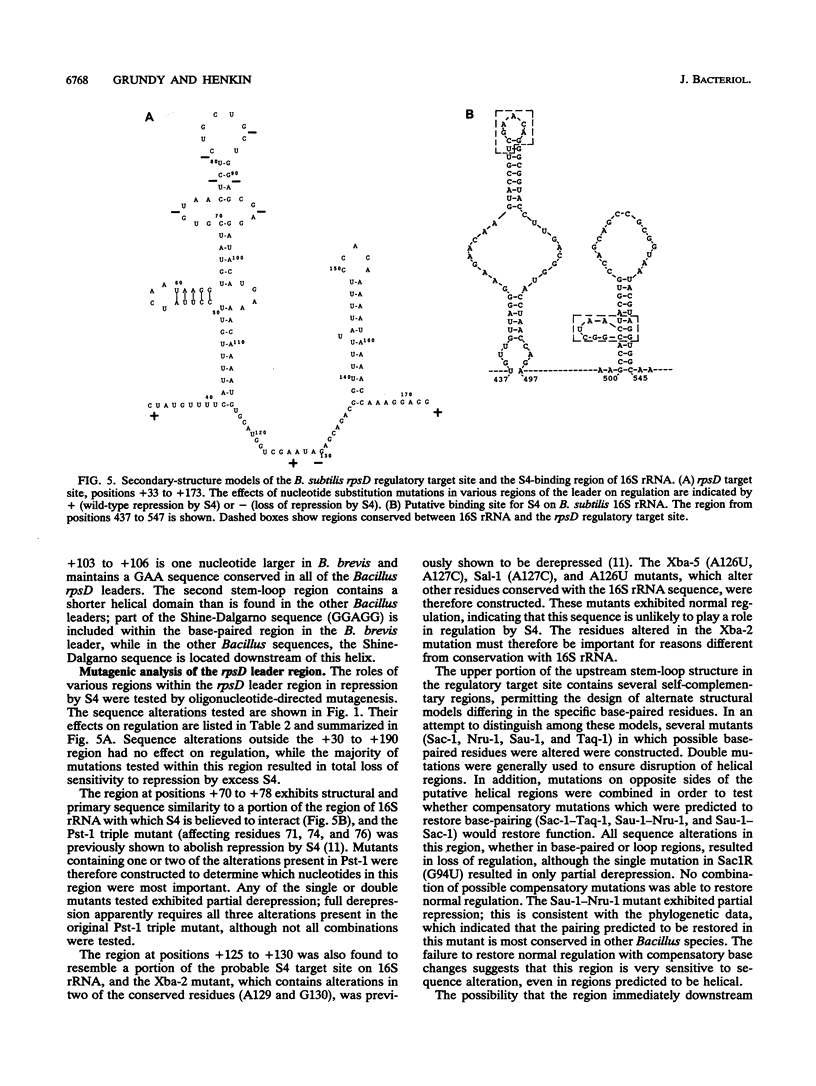
The Bacillus subtilis rpsD gene, which encodes ribosomal protein S4, is subject to autogenous regulation. Repression of rpsD expression by excess S4 protein was previously shown to be affected by mutations in the leader region of the gene. A large number of deletion and point mutations in the leader region were generated, and their effect on repression by S4 in vivo was tested. These studies indicated that the required region was within positions +30 to +190 relative to the transcription start point. Replacement of the rpsD promoter with a lac promoter derivative which is expressed in B. subtilis had no effect, indicating that repression by S4 occurs at a level subsequent to transcription initiation. The rpsD leader region was isolated from several Bacillus species. Members of the B. subtilis group, as defined by analysis of 16S rRNA sequence, contained a leader region target site very closely related in structure to that of B. subtilis, despite considerable primary sequence variation; the B. brevis rpsD leader contained some but not all of the structural features found in the regulatory target sites of the other Bacillus species. Very little similarity to the Escherichia coli alpha operon S4 target site was found at either the primary-sequence or the secondary-structure level. Mutagenic and phylogenetic data indicate that the secondary structure of the leader region regulatory target site contains two large stem-loop domains. The first of these helices has a side loop which is essential for autoregulation, is highly conserved among Bacillus rpsD genes, and is similar to a region of 16S rRNA important in S4 binding.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bedwell D., Davis G., Gosink M., Post L., Nomura M., Kestler H., Zengel J. M., Lindahl L. Nucleotide sequence of the alpha ribosomal protein operon of Escherichia coli. Nucleic Acids Res. 1985 Jun 11;13(11):3891–3903. doi: 10.1093/nar/13.11.3891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boylan S. A., Suh J. W., Thomas S. M., Price C. W. Gene encoding the alpha core subunit of Bacillus subtilis RNA polymerase is cotranscribed with the genes for initiation factor 1 and ribosomal proteins B, S13, S11, and L17. J Bacteriol. 1989 May;171(5):2553–2562. doi: 10.1128/jb.171.5.2553-2562.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Changchien L. M., Craven G. R. The function of the N-terminal region of ribosomal protein S4. J Mol Biol. 1976 Dec;108(2):381–401. doi: 10.1016/s0022-2836(76)80126-x. [DOI] [PubMed] [Google Scholar]
- Conrad R. C., Craven G. R. A cyanogen bromide fragment of S4 that specifically rebinds 16S RNA. Nucleic Acids Res. 1987 Dec 23;15(24):10331–10343. doi: 10.1093/nar/15.24.10331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deckman I. C., Draper D. E. Specific interaction between ribosomal protein S4 and the alpha operon messenger RNA. Biochemistry. 1985 Dec 31;24(27):7860–7865. doi: 10.1021/bi00348a002. [DOI] [PubMed] [Google Scholar]
- Green C. J., Stewart G. C., Hollis M. A., Vold B. S., Bott K. F. Nucleotide sequence of the Bacillus subtilis ribosomal RNA operon, rrnB. Gene. 1985;37(1-3):261–266. doi: 10.1016/0378-1119(85)90281-1. [DOI] [PubMed] [Google Scholar]
- Grundy F. J., Henkin T. M. Cloning and analysis of the Bacillus subtilis rpsD gene, encoding ribosomal protein S4. J Bacteriol. 1990 Nov;172(11):6372–6379. doi: 10.1128/jb.172.11.6372-6379.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundy F. J., Henkin T. M. The rpsD gene, encoding ribosomal protein S4, is autogenously regulated in Bacillus subtilis. J Bacteriol. 1991 Aug;173(15):4595–4602. doi: 10.1128/jb.173.15.4595-4602.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henkin T. M., Chambliss G. H. Genetic mapping of a mutation causing an alteration in Bacillus subtilis ribosomal protein S4. Mol Gen Genet. 1984;193(2):364–369. doi: 10.1007/BF00330694. [DOI] [PubMed] [Google Scholar]
- Henkin T. M., Sonenshein A. L. Mutations of the Escherichia coli lacUV5 promoter resulting in increased expression in Bacillus subtilis. Mol Gen Genet. 1987 Oct;209(3):467–474. doi: 10.1007/BF00331151. [DOI] [PubMed] [Google Scholar]
- Higo K. I., Loertscher K. Amino-terminal sequences of some Escherichia coli 30S ribosomal proteins and functionally corresponding Bacillus stearothermophilus ribosomal proteins. J Bacteriol. 1974 Apr;118(1):180–186. doi: 10.1128/jb.118.1.180-186.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higo K., Held W., Kahan L., Nomura M. Functional correspondence between 30S ribosomal proteins of Escherichia coli and Bacillus stearothermophilus. Proc Natl Acad Sci U S A. 1973 Mar;70(3):944–948. doi: 10.1073/pnas.70.3.944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinks-Robertson S., Nomura M. Ribosomal protein S4 acts in trans as a translational repressor to regulate expression of the alpha operon in Escherichia coli. J Bacteriol. 1982 Jul;151(1):193–202. doi: 10.1128/jb.151.1.193-202.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel T. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci U S A. 1985 Jan;82(2):488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messing J. New M13 vectors for cloning. Methods Enzymol. 1983;101:20–78. doi: 10.1016/0076-6879(83)01005-8. [DOI] [PubMed] [Google Scholar]
- Nomura M., Traub P., Bechmann H. Hybrid 30S ribosomal particles reconstituted from components of different bacterial origins. Nature. 1968 Aug 24;219(5156):793–799. doi: 10.1038/219793b0. [DOI] [PubMed] [Google Scholar]
- Rössler D., Ludwig W., Schleifer K. H., Lin C., McGill T. J., Wisotzkey J. D., Jurtshuk P., Jr, Fox G. E. Phylogenetic diversity in the genus Bacillus as seen by 16S rRNA sequencing studies. Syst Appl Microbiol. 1991;14(3):266–269. doi: 10.1016/S0723-2020(11)80379-6. [DOI] [PubMed] [Google Scholar]
- Stern S., Wilson R. C., Noller H. F. Localization of the binding site for protein S4 on 16 S ribosomal RNA by chemical and enzymatic probing and primer extension. J Mol Biol. 1986 Nov 5;192(1):101–110. doi: 10.1016/0022-2836(86)90467-5. [DOI] [PubMed] [Google Scholar]
- Tang C. K., Draper D. E. Evidence for allosteric coupling between the ribosome and repressor binding sites of a translationally regulated mRNA. Biochemistry. 1990 May 8;29(18):4434–4439. doi: 10.1021/bi00470a025. [DOI] [PubMed] [Google Scholar]
- Tang C. K., Draper D. E. Unusual mRNA pseudoknot structure is recognized by a protein translational repressor. Cell. 1989 May 19;57(4):531–536. doi: 10.1016/0092-8674(89)90123-2. [DOI] [PubMed] [Google Scholar]
- Thurlow D. L., Zimmermann R. A. Conservation of ribosomal protein binding sites in prokaryotic 16S RNAs. Proc Natl Acad Sci U S A. 1978 Jun;75(6):2859–2863. doi: 10.1073/pnas.75.6.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vartikar J. V., Draper D. E. S4-16 S ribosomal RNA complex. Binding constant measurements and specific recognition of a 460-nucleotide region. J Mol Biol. 1989 Sep 20;209(2):221–234. doi: 10.1016/0022-2836(89)90274-x. [DOI] [PubMed] [Google Scholar]
- Wei Y. G., Surzycki S. J. Screening recombinant clones containing sequences homologous to Escherichia coli genes using single-stranded bacteriophage vector. Gene. 1986;48(2-3):251–256. doi: 10.1016/0378-1119(86)90083-1. [DOI] [PubMed] [Google Scholar]
- Zuber P., Losick R. Role of AbrB in Spo0A- and Spo0B-dependent utilization of a sporulation promoter in Bacillus subtilis. J Bacteriol. 1987 May;169(5):2223–2230. doi: 10.1128/jb.169.5.2223-2230.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]



