Abstract
The E6 oncoprotein of bovine papillomavirus type 1 (BPV-1) has been shown to transform cells through a p53-independent pathway, but its transforming mechanism is unknown. Here we demonstrate in vitro and in vivo interactions between BPV-1 E6 and the focal adhesion protein paxillin. The ability of BPV-1 E6 to complex with paxillin correlated with its ability to transform; E6 mutant proteins impaired in their transformation function also were impaired in their abilities to bind paxillin. E6 binding to paxillin also may contribute to the carcinogenic potential of the human papillomavirus (HPV); we were able to show in vitro binding of paxillin to the E6 proteins of the cancer-associated type HPV 16 but not of the nononcogenic types 6 and 11. The association of E6 with paxillin was affected by depolymerization of the actin fiber network, and overexpression of BPV-1 E6 led to disruption of actin fiber formation. Disruption of the actin cytoskeleton is a characteristic of many transformed cells, and, in BPV-1 transformed cells, may be mediated by BPV-1 E6 through its interaction with paxillin.
Papillomaviruses are small DNA viruses that induce warts in a variety of animals, and certain types of human papillomaviruses (e.g., HPV16 and HPV18) have been associated with malignant epithelial tumors. The bovine papillomavirus type 1 (BPV-1) belongs to a subgroup of papillomaviruses that cause fibropapillomas in their natural hosts, with proliferation of both fibroblasts and squamous epithelial cells (1). BPV-1 has served as the prototype for the studies of various aspects of papillomavirus biology, including the mechanisms of cellular transformation. BPV-1 efficiently induces transformation of rodent cells in culture. Initial genetic studies mapped the BPV-1 transforming genes to two regions of the viral genome: the E5 ORF (2, 3) and the E6/E7 ORFs (4–6). Even though E5 or E6/E7 viral proteins can independently transform cells, they cooperate to give rise to high transformation efficiency (5, 7). The BPV-1 E5 transformation seems at least in part to be mediated through constitutive activation of growth factor receptors. It has been shown to directly bind and activate the β-receptor of the PDGF receptor (8, 9) and EGF receptor (10, 11).
In contrast, little is known about the transforming mechanism of BPV-1 E6. Although both E6 and E7 proteins are required for full transformation, there is evidence that E6 mutants are more defective in transformation than E7 mutants and that E6 but not E7 can independently transform mouse C127 cells (4, 7). The BPV-1 E6 gene product is a basic, 137-amino acid protein. It contains four Cys-X-X-Cys motifs that are conserved in all of the papillomavirus E6 proteins. Unlike the HPV16 and 18 E6, BPV-1 E6 does not bind to the tumor suppressor protein p53 (12) or stimulate E6AP-mediated ubiquitination and degradation (X.T., unpublished data). Therefore, BPV-1 E6 seems to induce cell transformation through a different pathway. BPV-1 E6 has been shown to bind in vitro to a 55-kDa putative calcium-binding protein (ERC-55) (13); however, the functional significance of this interaction has not yet been established.
In this report, we demonstrate interactions, both in vitro and in vivo, between BPV-1 E6 and the focal adhesion protein paxillin. Paxillin is a protein involved in transducing signals from the plasma membrane to focal adhesions and the actin cytoskeleton. Paxillin is tyrosine-phosphorylated in response to a variety of stimuli, including cross-linking of integrin molecules (14), treatment with growth factors (15, 16), and transformation by v-src (17), v-crk (18), or p210BCR/ABL (19). Tyrosine phosphorylation of focal adhesion proteins is closely associated with changes in the structure of the actin cytoskeleton although the precise downstream events of such phosphorylation are currently unknown. Paxillin has been shown to bind to β-integrin (20), oncoproteins such as v-Src (21), v-Crk (18), and p210BCR/ABL (19), and other focal adhesion proteins such as p125FAK (22), vinculin (23), and talin (19). Of interest, the high risk HPV 16 E6 oncoprotein but not the low risk HPV 6 or 11 E6 proteins also binds to paxillin. The actin cytoskeleton is altered in BPV-1 E6-transformed cells, and disruption of the actin cytoskeleton is demonstrated in cells acutely expressing BPV-1 E6. A model of how BPV-1 E5 and E6 may cooperate in cellular transformation is discussed.
MATERIALS AND METHODS
Immunoprecipitation of FLAG-Tagged E6.
The N-terminal, FLAG-tagged E6 was cloned into the pSG5–FLAG vector (24) by PCR. To make total cell lysates, COS-7 cells were labeled by [35S]cysteine and [35S]methionine and harvested 40 h after transfection and lysed in lysis buffer (20 mM Hepes/1% Nonidet P-40/150 mM NaCl/2 mM CaCl2/10% glycerol/1 mM phenylmethylsulfonyl fluoride/2 μg/ml aprotinin). Lysates were precipitated with the M2 mAb against the FLAG epitope (IBI). To purify p65, 50 × 150-mm dishes of COS cells were transfected with FLAG-tagged E6 and harvested as described above. Total cell lysates were purified by FLAG–antibody affinity chromatography (IBI). After extensive wash with lysis buffer plus 0.5 M NaCl, bound proteins were eluted by a 50-μg/ml FLAG peptide (IBI) and separated by SDS/PAGE. After transferring to poly(vinylidene difluoride) membrane (Bio-Rad), ≈3 μg of p65 was excised and subjected to microsequencing at the Harvard microchemistry facility (Cambridge, MA).
In Vitro Binding of E6 to Paxillin.
All E6 proteins were labeled with [35S]cysteine and methionine by in vitro translation (Promega) and incubated with full length glutathione S-transferase (GST)–paxillin fusion protein (amino acids 1–557) (19) in lysis buffer supplemented with 0.5 mg/ml BSA. Bound E6 proteins were analyzed by SDS/PAGE and autoradiography.
Cell Fractionation.
Cell fractionation was carried out as described (25) with the following modifications. Cells were washed once with cold PBS and lysed on plate in STM buffer (20 mM Hepes, pH 8.0/0.25 M sucrose/10 mM MgCl2/1 mM phenylmethylsulfonyl fluoride/2 μg/ml aprotinin/1 mM DTT). The cells were disrupted with 30 strokes in a glass Dounce homogenizer. Fraction a (cytosol) was prepared by centrifuging the lysates at 14,000 rpm for 20 min in a microcentrifuge. The membrane fraction (fraction b) was prepared by extracting the pellet from the spin with STM buffer plus 0.05% Nonidet P-40 and centrifuging at 14,000 rpm. The nuclear fraction (fraction c) was prepared by extracting the insoluble material from the membrane fraction with lysis buffer plus 0.1% SDS. All fractions were adjusted to lysis buffer plus 0.1% SDS before immunoprecipitation. Equal fractions of the immunoprecipitates were loaded in each lane. The Western blot was scanned and quantitated using National Institutes of Health image 1.55 software.
Immunofluorescence.
COS and C127 cells were transfected transiently with FLAG-tagged wild-type or mutant E6 constructs (H105D and Δ134–137) and harvested after 40 h. Cells were fixed with 4% paraformaldehyde and permeabilized with 0.5% Triton X-100. Cells were then blocked with 2.5% BSA plus 1% goat serum and doubled stained for F-actin using tetramethylrhodamine B isothiocyanate–phalloidin (Sigma) and for E6 using M2 antibody followed by fluorescein isothiocyanate-conjugated goat anti-mouse IgG (Sigma). C127 cells stably transfected with vector or transformed with E6 were stained for F-actin using tetramethylrhodamine B isothiocyanate–phalloidin as described above.
RESULTS
Identification of BPV-1-Associated Cellular Proteins.
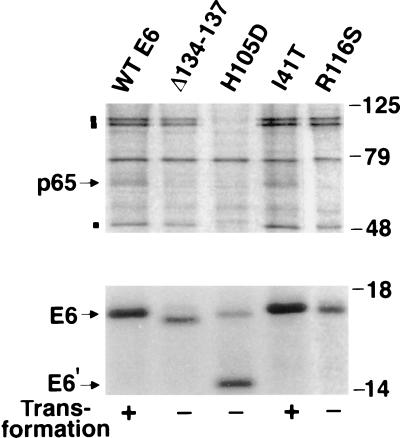
To identify cellular targets of BPV-E6 that might be relevant to its transformation capability, cellular proteins that could complex with E6 by coimmunoprecipitation were examined. For this purpose, we constructed FLAG epitope-tagged, wild-type E6 and E6 mutants that had been characterized for their ability to transform C127 cells (26). Mutation of the conserved CXXCs motif is likely to disrupt the overall structure of the E6 protein, so we focused on the mutants of amino acids other than cysteines (Fig. 1). The transforming function of each FLAG-tagged E6 protein was tested using C127 cells and was found to be the same as that of the untagged E6 protein (data not shown). COS cells were transfected transiently with FLAG-tagged, E6-encoding plasmids, and the E6 proteins were immunoprecipitated from total cell lysates of 35S-labeled cells using an antibody to the FLAG epitope. A cellular protein of 65 kDa (p65) specifically coprecipitated with transformation-competent E6 proteins (wild-type E6 and I41T) but not with transformation-deficient E6 mutants (Δ134–137, H105D, and R116S) (Fig. 1). Although additional cellular proteins of 110 kDa (p110), 100 kDa (p100), and 50 kDa (p50) were found to coprecipitate with wild-type E6, their binding specificity did not correlate with the E6-transforming activity; two nontransforming E6 mutants (Δ134–137, R116S) also bound to p110, p100, and p50 (Fig. 1). Our major interest was in cellular proteins that might be implicated in E6 transformation, so we therefore focused our initial work on the identification of p65. Approximately 3 μg of p65 was subsequently purified by FLAG antibody affinity chromatography for microsequencing. The amino acid sequence of two p65 peptides was obtained that matched the sequence of the focal adhesion protein paxillin (amino acids 394–418 and 433–440, respectively).
Figure 1.
Identification of BPV-1 E6-associated protein. COS cells transfected with FLAG-tagged wild-type E6 or mutant E6 DNAs were 35S-labeled, and total cell lysates were immunoprecipitated with M2 FLAG antibody. The immunoprecipitates were analyzed by SDS/PAGE and autoradiography. E6′ is probably a degradation product. The positions of p65 and E6 are indicated by arrows and the positions of p110, p100, and p50 are indicated by ▪.
In Vitro Interaction Between Paxillin and BPV-1 E6 Proteins.
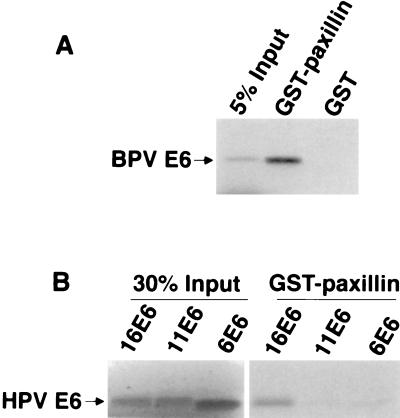
To verify the interaction between paxillin and BPV-1 E6, a GST–paxillin fusion protein was used to bind to 35S-labeled, in vitro-translated BPV-1 E6. As shown in Fig. 2A, GST–paxillin bound to ≈30% of input BPV-E6 whereas the control GST protein had no detectable binding.
Figure 2.
(A) In vitro interaction between BPV-1 E6 and paxillin. GST–paxillin fusion protein (19) and GST protein were incubated with 35S-labeled, in vitro-translated BPV-1 E6, and bound E6 proteins were analyzed by SDS/PAGE and autoradiography. (B) Interaction between paxillin and HPV E6 proteins. HPV 16, 11, and 6 E6 proteins were 35S-labeled by in vitro translation and incubated with GST–paxillin, and bound E6 proteins were analyzed by SDS/PAGE and autoradiography.
To further assess the correlation between the ability of BPV-1 E6 to bind to paxillin and its ability to transform C127 cells, a panel of E6 mutants was tested in the GST–paxillin binding assay. As shown in Table 1, E6 proteins with wild-type transforming activity bound to GST–paxillin with near wild-type efficiency whereas nontransforming E6 mutant proteins were defective in paxillin binding or, in the case of three mutants (H105D, R116S, and Δ134–137), bound to paxillin with about 10–20% of the efficiency of the wild-type E6 protein. None of these three mutants, however, coimmunoprecipitated with paxillin in vivo (Fig. 1; Table 1). Therefore, the combination of in vitro and in vivo binding experiments demonstrated a good correlation between the ability of BPV-1 E6 to transform and to efficiently bind paxillin.
Table 1.
Paxillin binding to BPV-1 E6 mutants
| E6 mutant | Foci, n* | Growth in agar* | Paxillin binding
|
|
|---|---|---|---|---|
| In vitro† | In vivo‡ | |||
| Wild-type | 68 | + | 100 | + |
| I41T | 64 | + | 81 | + |
| C128S | 26 | + | 93 | ND |
| C17P | 0 | − | 0 | ND |
| C20S | 0 | − | 0 | ND |
| C53R | 0 | − | 0 | ND |
| C90S | 0 | − | 0 | ND |
| C93H | 0 | − | 0 | ND |
| C93S | 0 | − | 0 | ND |
| H105D | 0 | − | 12 | − |
| R116S | 0 | − | 19 | − |
| C124V | 0 | − | 0 | ND |
| Δ127-137 | 0 | − | 0 | ND |
| Δ134-137 | 0 | − | 18 | − |
ND, not determined.
Data are derived from Vousden et al. (26).
In vitro binding was determined by binding of in vitro-translated E6 products to GST–paxillin.
In vivo binding was determined by coimmunoprecipitation of paxillin and FLAG-tagged E6 in COS cells.
In Vitro Interaction Between Paxillin and HPV E6 Proteins.
Although the E6 proteins of the cancer-associated high risk HPVs have been shown to functionally inactivate p53 (27) and to mediate its ubiquitination and degradation (28–30), there is evidence that HPV 16 E6 protein may possess p53-independent transforming functions. Mutational analysis has suggested that E6-mediated degradation of p53 is necessary but not sufficient for transformation of human embryonic kidney cells (31). Furthermore, HPV 16 E6 activates telomerase in a p53-independent manner in human keratinocytes and mammary epithelial cells (32). To investigate whether paxillin could be involved in HPV E6 transformation, we extended our analysis and tested whether HPV E6 proteins could bind to paxillin. High risk HPV 16 E6 and low risk HPV 11 and HPV 6 E6 proteins were 35S-labeled by in vitro translation and tested for their binding with GST–paxillin. As shown in Fig. 2B, GST–paxillin bound to ≈20% of the high risk HPV16 E6 but not to the low risk 11 E6 and 6 E6 proteins. Low risk HPV 6 and HPV 11 E6 proteins have not been shown to have transformation properties, so these results provide additional correlation between paxillin–E6 binding and cellular transformation.
The Effect of Cytochalasin D on Paxillin–E6 Interaction.
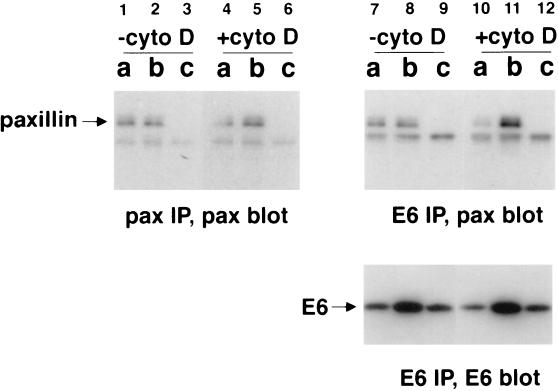
To further study the in vivo interaction between paxillin and BPV-1 E6, COS cells were transfected transiently with FLAG-tagged E6 and fractionated into a cytosolic fraction (fraction a), a membrane fraction (fraction b), and a nuclear fraction (fraction c) (25) for immunoprecipitation and Western blot analysis. Paxillin is a component of focal adhesions and the cytoskeleton, so cells also were treated with cytochalasin D, an actin-polymerization inhibitor, to determine whether the paxillin–E6 interaction would be affected by disruption of the actin cytoskeleton. As controls for the efficiency of fractionation, lysates from each fraction were blotted for a nuclear protein (simian virus 40 T antigen), a membrane protein (transferrin receptor), and a cytoplasmic protein (lactate dehydrogenase) and were found to contain the appropriate marker protein (data not shown). The fractionation experiment revealed that paxillin was present in the cytosolic and membrane fractions. The cytoskeleton-associated paxillin was expected to be fractionated in the membrane fraction (Fig. 3, lanes 1–3). The distribution of BPV-1 E6 was similar to what has been reported in E6-transformed C127 cells (25) (Fig. 3, lanes 7–9, bottom) although we did observe an increased amount of cytosolic E6 presumably due to the higher levels of E6 expression achieved in COS cells. Important to note, paxillin was coprecipitated with E6 using the FLAG antibody (Fig. 3, lanes 7–9, top), further demonstrating that the interaction between paxillin and E6 occurs in vivo. In these experiments, because of the relative inefficiency of the paxillin antibody in immunoprecipitation, almost equal amounts of paxillin were precipitated by the paxillin antibody directly and by the FLAG antibody that coprecipitated E6 and paxillin (Fig. 3, compare lanes 1 and 2 to lanes 7 and 8, top). The interaction with paxillin was specific for wild-type E6 because immunoprecipitation of the nontransforming E6 mutant (H105D) did not bring down paxillin in this assay (data not shown). Upon treatment of cytochalasin D, a small amount of paxillin (15%) and E6 (12%) shifted from the cytosolic fraction to the membrane fraction, consistent with the observation that cytochalasin D causes redistributing of actin-containing cytoskeletal assemblies (33) (Fig. 3, compare lanes 1–2 to lanes 4–5 and lanes 7–8 to lanes 10–11, bottom). Surprisingly, cytochalasin D caused a much larger change in the amount of E6-associated paxillin. In the cytosolic fraction, the amount of paxillin precipitated by E6 was decreased by 40%; in the membrane fraction, the amount of paxillin precipitated by E6 was increased by 74% (Fig. 3, compare lanes 7 and 8 to lanes 10 and 11, top). These data indicate that the interaction between E6 and paxillin is sensitive to the status of actin cytoskeleton. In addition to causing the modest redistribution of paxillin and E6, cytochalasin D seemed to enhance the paxillin–E6 interaction in the membrane fraction while decreasing it in the cytosolic fraction.
Figure 3.
Study of E6–paxillin interaction by cell fractionation and cytochalasin D treatment. COS cells transfected with FLAG-tagged E6 were fractionated into cytosolic (fraction a), membrane (fraction b), and nuclear (fraction c) fractions. Half of the cells were treated with 2 μM cytochalasin D (Sigma) for 1 h before harvest. Fractionated lysates were immunoprecipitated using a paxillin mAb (Chemicon) or the M2 FLAG antibody and blotted for paxillin or FLAG–E6 as indicated.
The Effect of BPV-1 E6 on Paxillin and Actin Cytoskeleton.
Tyrosine phosphorylation of paxillin seems to be the key in the regulation of actin cytoskeleton organization, so we next studied whether BPV-1 E6 affected paxillin tyrosine phosphorylation. COS cells transiently transfected with BPV-1 E6 or C127 cells stably transformed by BPV-1 E6 were immunoprecipitated for paxillin and blotted using paxillin antibody or anti-P-tyr antibody. In these experiments, we observed no consistent change in the steady-state level of paxillin or its tyrosine phosphorylation comparing BPV-1 E6-expressing cells to vector-transfected cells (data not shown). Therefore, unlike other paxillin-binding oncoproteins such as v-src, v-crk, and p210BCR/ABL, which drastically increase paxillin tyrosine phosphorylation, E6 has no obvious effect on its tyrosine phosphorylation status, suggesting that E6 may interfere paxillin function through a novel pathway.
Transformed cells commonly exhibit altered morphology and reduced cell adherence due to disruption of cytoskeletal structures, so we investigated the possibility that E6 may be directly involved in such processes, possibly through its interaction with paxillin. We therefore studied the effect of BPV-1 E6 expression on paxillin subcellular localization and actin fiber distribution. COS cells transiently expressing E6 were double-stained for paxillin and for E6. In both E6-positive and E6-negative cells, paxillin was found to localize to focal adhesion-like structures as reported (17, 34), and there were no gross morphological changes in focal adhesions in E6-expressing cells (data not shown). However, we cannot exclude the possibility that E6 may have subtle effects on the organization of focal adhesions that could not be detected by the paxillin antibody used in these studies. Similar to the results obtained with COS cells, C127 cells that were transiently transfected with E6 or that were stably transformed by E6 each exhibited normal paxillin localization (data not shown).
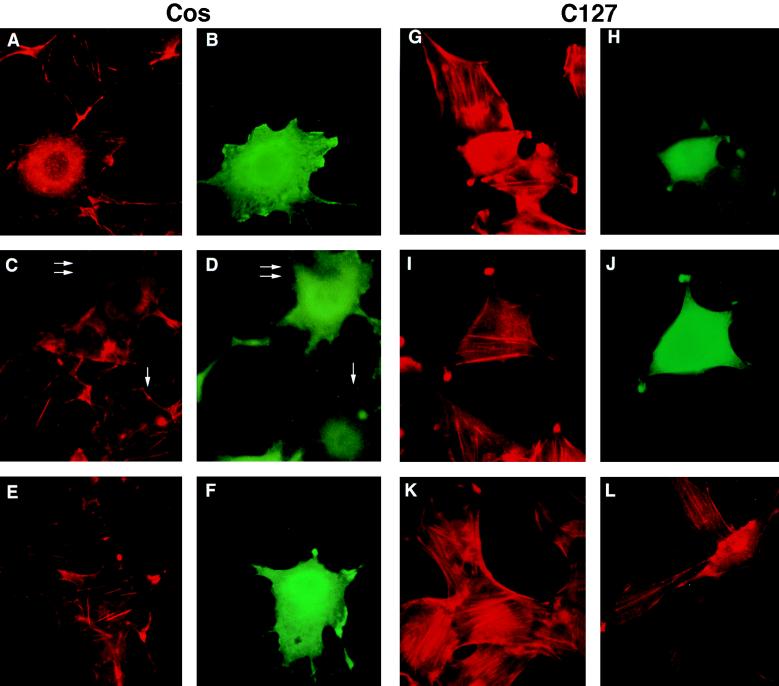
We next examined the effect of BPV-1 E6 expression on the actin fiber network. COS cells transfected with FLAG-tagged E6 were double-stained for E6 by the FLAG antibody and F-actin by phalloidin. In COS cells overexpressing wild-type E6, actin fibers were disrupted (Fig. 4, A and B). Furthermore, the disruption of actin cables was dependent on the E6 expression level because some actin fibers were evident in cells with low E6 signal (Fig. 4, C and D, compare the cell with low E6 expression indicated by a single arrow to the cell with high E6 level indicated by a double arrow). Disruption of actin fiber was observed in 54% of COS cells with high E6 levels (84 cells examined) whereas only 6% of nontransfected cells (140 cells examined) did not have actin fibers. In addition, the E6 disruption of actin fiber correlated with its ability to bind to paxillin and to transform. The nontransforming mutant E6 proteins (Δ134–137, H105D) that did not bind to paxillin failed to disrupt the actin fibers in COS cells (Fig. 4 E and F and data not shown). Similarly, wild-type E6 but not mutant E6 disrupted actin fibers in transiently transfected C127 cells although there was more diffuse F-actin staining in E6-containing C127 cells than in COS cells (Fig. 4, G-J), probably because of more abundant actin fibers in C127 cells. In C127 cells stably transformed by E6, the phalloidin staining revealed only a few fine actin fibers (Fig. 4L) whereas nontransformed vector-transfected C127 cells contained multiple heavy actin cables (Fig. 4K). The presence of the thin actin fibers in E6-transformed C127 cells compared with the complete lack of actin fiber in E6-transfected cells was likely caused by the much lower E6 level in stably transformed cells compared with transiently transfected cells. Concomitant with the alteration in the actin stress fiber in E6-transformed C127 cells, there was a marked change in cell morphology in that the E6-expressing C127 cells were spindled-shaped whereas the nontransformed vector- containing cells remained flat, suggesting that disruption of the actin cytoskeleton structure by E6 may contribute to the altered cell shape of the transformed cells.
Figure 4.
Disruption of actin fiber by BPV-1 E6 expression. COS cells or C127 cells were transfected with FLAG-tagged wild-type E6 (A–D, G, and H) or mutant E6 (H105D; E, F, I, and J) and double-stained for F-actin using tetramethylrhodamine B isothiocyanate–phalloidin (A, C, E, G, and I) and for E6 using M2 antibody (B, D, F, H, and J). In C and D, the double arrow indicated a cell with strong E6 signal, and the single arrow indicated a cell with a weak E6 signal. C127 cells stably transfected with vector (K) or transformed by E6 (L) were stained for F-actin using tetramethylrhodamine B isothiocyanate–phalloidin.
DISCUSSION
In conclusion, the data suggest that the BPV-1 E6 oncoprotein disrupts actin fiber formation through its interaction with the focal adhesion protein paxillin. The mechanism of how this change may occur is yet unclear. The observation that BPV-1 E6 has no effect on tyrosine phosphorylation of paxillin suggests that the effect of E6 on paxillin is downstream of paxillin tyrosine phosphorylation. It is also interesting that the E6 effect on actin network is E6 dose-dependent in transient expression experiments in that higher E6 levels result in more severe actin fiber disruption, suggesting that E6 is the rate-limiting factor in this process. In addition, the E6–paxillin association appears to be sensitive to the status of actin polymerization. Depolymerization of actin fibers by cytochalasin D results in a slight shift of paxillin and E6 from the cytosol to the cytoskeleton and also affects the affinity of the E6–paxillin interaction, suggesting a possible feedback mechanism from the actin cytoskeleton to paxillin and E6. The correlation between paxillin binding and transforming competence for both HPV and BPV-1 E6 proteins suggests an important role for the E6–paxillin interaction in the oncogenic process.
The actin cytoskeleton is critical for many aspects of cell function. In addition to maintaining cell morphology, it is required for cell motility, cell division, cell–cell contact and cell–extracellular matrix contact (35). Consistent with the proposed role for E6 in organizing the actin cytoskeleton, E6 has been shown to be required for the anchorage-independent growth of BPV-1-transformed cells (7, 36). Therefore, the E6 interference with the actin fiber network could result in profound changes in cellular signal transduction and cell cycle control, thereby contributing to the transformed cell phenotype.
Our data also shed light on how the BPV-1 E5 and E6 oncoproteins may cooperate in transformation. Normal cells require both adhesion to extracellular matrix proteins and stimulation by serum or growth factors to proliferate. These requirements reflect the fact that adhesion molecules and growth factor receptors transmit signals that eventually converge to determine cell cycle progression. In contrast, one characteristic of transformed cells is independence from cellular controls mediated by both adhesion and serum (37, 38). The observations that BPV-1 E5 activates the platelet-derived growth factor and epidermal growth factor receptor and that E6 disrupts paxillin function as shown in this report support the view that signals from cell adhesion and growth factor receptor are both required for regulated cell proliferation and that disruption of these pathways cooperatively contribute to tumorigenesis.
Acknowledgments
We thank R. Salgia, J. L. Li, and J. Griffin for providing GST–paxillin, E. Hatzivassiliou for providing the pSG5–FLAG vector, and J. Chen and E. J. Androphy for providing BPV-1 E6 mutants. We also thank J. Griffin for helpful discussions and for critically reading the manuscript. X.T. was supported by the cancer research fund of the Damon Runyon–Walter Winchell Foundation; this research was supported by National Institutes of Health Grant P01CA50661-08 (PH).
ABBREVIATIONS
- BPV-1
bovine papillomavirus type 1
- HPV
human papillomavirus
- GST
glutathione S-transferase
References
- 1.Howley P M. In: Virology. Fields B N, Howley P M, Knipe D M, editors. New York: Raven; 1995. pp. 2045–2076. [Google Scholar]
- 2.Groff D E, Lancaster W D. Virology. 1986;150:221–230. doi: 10.1016/0042-6822(86)90281-3. [DOI] [PubMed] [Google Scholar]
- 3.Schiller J T, Vass W C, Vousden K H, Lowy D R. J Virol. 1986;57:1–6. doi: 10.1128/jvi.57.1.1-6.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schiller J T, Vass W C, Lowy D R. Proc Natl Acad Sci USA. 1984;81:7880–7884. doi: 10.1073/pnas.81.24.7880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang Y-C, Okayama H, Howley P M. Proc Natl Acad Sci USA. 1985;82:1030–1034. doi: 10.1073/pnas.82.4.1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vande Pol S B, Howley P M. J Virol. 1995;69:395–402. doi: 10.1128/jvi.69.1.395-402.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Neary K, DiMaio D. J Virol. 1989;63:259–266. doi: 10.1128/jvi.63.1.259-266.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goldstein D J, Andresson T, Sparkowski J J, Schlegel R. EMBO J. 1992;11:4851–4859. doi: 10.1002/j.1460-2075.1992.tb05591.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Petti L, DiMaio D. Proc Natl Acad Sci USA. 1992;89:6736–6740. doi: 10.1073/pnas.89.15.6736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martin P, Vass W C, Schiller J T, Lowy D R, Velu T J. Cell. 1989;59:21–32. doi: 10.1016/0092-8674(89)90866-0. [DOI] [PubMed] [Google Scholar]
- 11.Cohen B D, Goldstein D J, Rutledge L, Vass W C, Lowy D R, Schlegel R, Schiller J T. J Virol. 1993;67:5303–5311. doi: 10.1128/jvi.67.9.5303-5311.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Werness B A, Levine A J, Howley P M. Science. 1990;248:76–79. doi: 10.1126/science.2157286. [DOI] [PubMed] [Google Scholar]
- 13.Chen J J, Reid C E, Band V, Androphy E J. Science. 1995;269:529–531. doi: 10.1126/science.7624774. [DOI] [PubMed] [Google Scholar]
- 14.Burridge K, Turner C E, Romer L H. J Cell Biol. 1992;119:893–903. doi: 10.1083/jcb.119.4.893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Khan M A, Okumura N, Okada M, Kobayashi S, Nakagawa H. FEBS Lett. 1995;362:201–204. doi: 10.1016/0014-5793(95)00250-d. [DOI] [PubMed] [Google Scholar]
- 16.Rankin S, Rozengurt E. J Biol Chem. 1994;269:704–710. [PubMed] [Google Scholar]
- 17.Glenney J R, Zokas L. J Cell Biol. 1989;108:2401–2408. doi: 10.1083/jcb.108.6.2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Birge R B, Fajardo J E, Reichman C, Shoelson S E, Zhou S, Cantley L C, Hanafusa H. Mol Cell Biol. 1993;13:4648–4656. doi: 10.1128/mcb.13.8.4648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Salgia R, Li J L, Lo S H, Brunkhorst B, Kanas G S, Sobhany E S, Pisick E, Hallek M, Ernst T, Tantravahi R, Chen L B, Griffin J D. J Biol Chem. 1995;270:5039–5047. doi: 10.1074/jbc.270.10.5039. [DOI] [PubMed] [Google Scholar]
- 20.Schaller M D, Otey C A, Hildebrand J D, Parsons J T. J Cell Biol. 1995;130:1181–1187. doi: 10.1083/jcb.130.5.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weng Z, Taylor J A, Turner C E, Brugge J S, Seidel-Dugan C. J Biol Chem. 1993;268:14956–14963. [PubMed] [Google Scholar]
- 22.Tachibana K, Sato T, D’Avirro N, Morimoto C. J Exp Med. 1995;182:1089–1100. doi: 10.1084/jem.182.4.1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wood C K, Turner C E, Jackson P, Critchley D R. J Cell Sci. 1994;107:709–717. [PubMed] [Google Scholar]
- 24.Hatzivassiliou, E., Cardot, P., Zannis, V. & Mitsialis, A. (1997) Biochemistry, in press. [DOI] [PubMed]
- 25.Androphy E J, Schiller J T, Lowy D R. Science. 1985;230:442–445. doi: 10.1126/science.2996134. [DOI] [PubMed] [Google Scholar]
- 26.Vousden K H, Androphy E J, Schiller J T, Lowy D R. J Virol. 1989;63:2340–2342. doi: 10.1128/jvi.63.5.2340-2342.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mietz J A, Unger T, Huibregtse J M, Howley P M. EMBO J. 1992;11:5013–5020. doi: 10.1002/j.1460-2075.1992.tb05608.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Scheffner M, Werness B A, Huibregtse J M, Howley P M. Cell. 1990;63:1129–1136. doi: 10.1016/0092-8674(90)90409-8. [DOI] [PubMed] [Google Scholar]
- 29.Scheffner M, Huibregtse J M, Vierstra R D, Howley P M. Cell. 1993;75:495–505. doi: 10.1016/0092-8674(93)90384-3. [DOI] [PubMed] [Google Scholar]
- 30.Huibregtse J M, Scheffner M, Howley P M. Mol Cell Biol. 1993;13:775–784. doi: 10.1128/mcb.13.2.775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nakagawa S, Watanabe S, Yoshikawa H, Taketani Y, Yoshiike K, Kanda T. Virology. 1995;212:535–542. doi: 10.1006/viro.1995.1511. [DOI] [PubMed] [Google Scholar]
- 32.Klingelhutz A J, Foster S A, McDougall J K. Nature (London) 1996;380:79–82. doi: 10.1038/380079a0. [DOI] [PubMed] [Google Scholar]
- 33.Brett J G, Godman G C. Tissue Cell. 1986;18:175–199. doi: 10.1016/0040-8166(86)90027-3. [DOI] [PubMed] [Google Scholar]
- 34.Turner C E, Glenney J R, Burridge K. J Cell Biol. 1990;111:1059–1068. doi: 10.1083/jcb.111.3.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ridley A J. Curr Opin Genet Dev. 1995;5:24–30. doi: 10.1016/s0959-437x(95)90049-7. [DOI] [PubMed] [Google Scholar]
- 36.Sarver N, Rabson M S, Yang Y C, Byrne J C, Howley P M. J Virol. 1984;52:377–388. doi: 10.1128/jvi.52.2.377-388.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schwartz M A, Schaller M D, Ginsberg M H. Annu Rev Cell Biol. 1995;11:549–599. doi: 10.1146/annurev.cb.11.110195.003001. [DOI] [PubMed] [Google Scholar]
- 38.Schwartz M A, Toksoz D, Khosravi-Far R. EMBO J. 1996;15:6525–6530. [PMC free article] [PubMed] [Google Scholar]