Abstract
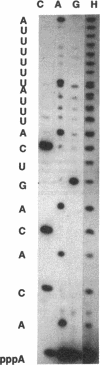
The Escherichia coli araFGH operon codes for proteins involved in the L-arabinose high-affinity transport system. Transcriptional regulation of the operon was studied by creating point mutations and deletions in the control region cloned into a GalK expression vector. The transcription start site was confirmed by RNA sequencing of transcripts. The sequences essential for polymerase function were localized by deletions and point mutations. Surprisingly, only a weak -10 consensus sequence, and no -35 sequence is required. Mutation of a guanosine at position -12 greatly reduced promoter activity, which suggests important polymerase interactions with DNA between the usual -10 and -35 positions. A double mutation toward the consensus in the -10 region was required to create a promoter capable of significant AraC-independent transcription. These results show that the araFGH promoter structure is similar to that of the galP1 promoter and is substantially different from that of the araBAD promoter. The effects of 11 mutations within the DNA region thought to bind the cyclic AMP receptor protein correlate well with the CRP consensus binding sequence and confirm that this region is responsible for cyclic AMP regulation. Deletion of the AraC binding site nearest the promoter, araFG1, eliminates arabinose regulation, whereas deletion of the upstream AraC binding site, araFG2, has only a slight effect on promoter activity.
Full text
PDF









Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adams C. W., Hatfield G. W. Effects of promoter strengths and growth conditions on copy number of transcription-fusion vectors. J Biol Chem. 1984 Jun 25;259(12):7399–7403. [PubMed] [Google Scholar]
- Auble D. T., deHaseth P. L. Promoter recognition by Escherichia coli RNA polymerase. Influence of DNA structure in the spacer separating the -10 and -35 regions. J Mol Biol. 1988 Aug 5;202(3):471–482. doi: 10.1016/0022-2836(88)90279-3. [DOI] [PubMed] [Google Scholar]
- Ayers D. G., Auble D. T., deHaseth P. L. Promoter recognition by Escherichia coli RNA polymerase. Role of the spacer DNA in functional complex formation. J Mol Biol. 1989 Jun 20;207(4):749–756. doi: 10.1016/0022-2836(89)90241-6. [DOI] [PubMed] [Google Scholar]
- Berg O. G., von Hippel P. H. Selection of DNA binding sites by regulatory proteins. II. The binding specificity of cyclic AMP receptor protein to recognition sites. J Mol Biol. 1988 Apr 20;200(4):709–723. doi: 10.1016/0022-2836(88)90482-2. [DOI] [PubMed] [Google Scholar]
- Berg O. G., von Hippel P. H. Selection of DNA binding sites by regulatory proteins. Statistical-mechanical theory and application to operators and promoters. J Mol Biol. 1987 Feb 20;193(4):723–750. doi: 10.1016/0022-2836(87)90354-8. [DOI] [PubMed] [Google Scholar]
- Brown C. E., Hogg R. W. A second transport system for L-arabinose in Escherichia coli B-r controlled by the araC gene. J Bacteriol. 1972 Aug;111(2):606–613. doi: 10.1128/jb.111.2.606-613.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busby S., Dreyfus M. Segment-specific mutagenesis of the regulatory region in the Escherichia coli galactose operon: isolation of mutations reducing the initiation of transcription and translation. Gene. 1983 Jan-Feb;21(1-2):121–131. doi: 10.1016/0378-1119(83)90154-3. [DOI] [PubMed] [Google Scholar]
- Chan B., Busby S. Recognition of nucleotide sequences at the Escherichia coli galactose operon P1 promoter by RNA polymerase. Gene. 1989 Dec 14;84(2):227–236. doi: 10.1016/0378-1119(89)90496-4. [DOI] [PubMed] [Google Scholar]
- Clark A. F., Hogg R. W. High-affinity arabinose transport mutants of Escherichia coli: isolation and gene location. J Bacteriol. 1981 Sep;147(3):920–924. doi: 10.1128/jb.147.3.920-924.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colomé J., Wilcox G., Englesberg E. Constitutive mutations in the controlling site region of the araBAD operon of Escherichia coli B/r that decrease sensitivity to catabolite repression. J Bacteriol. 1977 Feb;129(2):948–958. doi: 10.1128/jb.129.2.948-958.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dandanell G., Hammer K. deoP1 promoter and operator mutants in Escherichia coli: isolation and characterization. Mol Microbiol. 1991 Oct;5(10):2371–2376. doi: 10.1111/j.1365-2958.1991.tb02083.x. [DOI] [PubMed] [Google Scholar]
- Dunn T. M., Schleif R. Deletion analysis of the Escherichia coli ara PC and PBAD promoters. J Mol Biol. 1984 Nov 25;180(1):201–204. doi: 10.1016/0022-2836(84)90437-6. [DOI] [PubMed] [Google Scholar]
- Ebright R. H., Cossart P., Gicquel-Sanzey B., Beckwith J. Mutations that alter the DNA sequence specificity of the catabolite gene activator protein of E. coli. Nature. 1984 Sep 20;311(5983):232–235. doi: 10.1038/311232a0. [DOI] [PubMed] [Google Scholar]
- Ebright R. H., Ebright Y. W., Gunasekera A. Consensus DNA site for the Escherichia coli catabolite gene activator protein (CAP): CAP exhibits a 450-fold higher affinity for the consensus DNA site than for the E. coli lac DNA site. Nucleic Acids Res. 1989 Dec 25;17(24):10295–10305. doi: 10.1093/nar/17.24.10295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Englesberg E., Irr J., Power J., Lee N. Positive control of enzyme synthesis by gene C in the L-arabinose system. J Bacteriol. 1965 Oct;90(4):946–957. doi: 10.1128/jb.90.4.946-957.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eschenlauer A. C., Reznikoff W. S. Escherichia coli catabolite gene activator protein mutants defective in positive control of lac operon transcription. J Bacteriol. 1991 Aug;173(16):5024–5029. doi: 10.1128/jb.173.16.5024-5029.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaal T., Barkei J., Dickson R. R., deBoer H. A., deHaseth P. L., Alavi H., Gourse R. L. Saturation mutagenesis of an Escherichia coli rRNA promoter and initial characterization of promoter variants. J Bacteriol. 1989 Sep;171(9):4852–4861. doi: 10.1128/jb.171.9.4852-4861.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardella T., Moyle H., Susskind M. M. A mutant Escherichia coli sigma 70 subunit of RNA polymerase with altered promoter specificity. J Mol Biol. 1989 Apr 20;206(4):579–590. doi: 10.1016/0022-2836(89)90567-6. [DOI] [PubMed] [Google Scholar]
- Gaston K., Bell A., Kolb A., Buc H., Busby S. Stringent spacing requirements for transcription activation by CRP. Cell. 1990 Aug 24;62(4):733–743. doi: 10.1016/0092-8674(90)90118-x. [DOI] [PubMed] [Google Scholar]
- Gielow L., Largen M., Englesberg E. Initiator constitutive mutants of the L-arabinose operon (OIBAD) of Escherichia coli B/r. Genetics. 1971 Nov;69(3):289–302. doi: 10.1093/genetics/69.3.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graña D., Gardella T., Susskind M. M. The effects of mutations in the ant promoter of phage P22 depend on context. Genetics. 1988 Oct;120(2):319–327. doi: 10.1093/genetics/120.2.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenblatt J., Schleif R. Arabinose C protein: regulation of the arabinose operon in vitro. Nat New Biol. 1971 Oct 6;233(40):166–170. doi: 10.1038/newbio233166a0. [DOI] [PubMed] [Google Scholar]
- Hahn S., Dunn T., Schleif R. Upstream repression and CRP stimulation of the Escherichia coli L-arabinose operon. J Mol Biol. 1984 Nov 25;180(1):61–72. doi: 10.1016/0022-2836(84)90430-3. [DOI] [PubMed] [Google Scholar]
- Hahn S., Hendrickson W., Schleif R. Transcription of Escherichia coli ara in vitro. The cyclic AMP receptor protein requirement for PBAD induction that depends on the presence and orientation of the araO2 site. J Mol Biol. 1986 Apr 5;188(3):355–367. doi: 10.1016/0022-2836(86)90160-9. [DOI] [PubMed] [Google Scholar]
- Harley C. B., Reynolds R. P. Analysis of E. coli promoter sequences. Nucleic Acids Res. 1987 Mar 11;15(5):2343–2361. doi: 10.1093/nar/15.5.2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawley D. K., McClure W. R. Compilation and analysis of Escherichia coli promoter DNA sequences. Nucleic Acids Res. 1983 Apr 25;11(8):2237–2255. doi: 10.1093/nar/11.8.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickson W., Rudd K. E. Physical map location of the argFGH operon of Escherichia coli. J Bacteriol. 1992 Jun;174(11):3836–3837. doi: 10.1128/jb.174.11.3836-3837.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickson W., Schleif R. F. Regulation of the Escherichia coli L-arabinose operon studied by gel electrophoresis DNA binding assay. J Mol Biol. 1984 Sep 25;178(3):611–628. doi: 10.1016/0022-2836(84)90241-9. [DOI] [PubMed] [Google Scholar]
- Hendrickson W., Stoner C., Schleif R. Characterization of the Escherichia coli araFGH and araJ promoters. J Mol Biol. 1990 Oct 20;215(4):497–510. doi: 10.1016/S0022-2836(05)80163-9. [DOI] [PubMed] [Google Scholar]
- Hogg R. W., Englesberg E. L-arabinose binding protein from Escherichia coli B-r. J Bacteriol. 1969 Oct;100(1):423–432. doi: 10.1128/jb.100.1.423-432.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horazdovsky B. F., Hogg R. W. High-affinity L-arabinose transport operon. Gene product expression and mRNAs. J Mol Biol. 1987 Sep 5;197(1):27–35. doi: 10.1016/0022-2836(87)90606-1. [DOI] [PubMed] [Google Scholar]
- Horwitz A. H., Morandi C., Wilcox G. Deoxyribonucleic acid sequence of araBAD promoter mutants of Escherichia coli. J Bacteriol. 1980 May;142(2):659–667. doi: 10.1128/jb.142.2.659-667.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huo L., Martin K. J., Schleif R. Alternative DNA loops regulate the arabinose operon in Escherichia coli. Proc Natl Acad Sci U S A. 1988 Aug;85(15):5444–5448. doi: 10.1073/pnas.85.15.5444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kammerer W., Deuschle U., Gentz R., Bujard H. Functional dissection of Escherichia coli promoters: information in the transcribed region is involved in late steps of the overall process. EMBO J. 1986 Nov;5(11):2995–3000. doi: 10.1002/j.1460-2075.1986.tb04597.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keilty S., Rosenberg M. Constitutive function of a positively regulated promoter reveals new sequences essential for activity. J Biol Chem. 1987 May 5;262(13):6389–6395. [PubMed] [Google Scholar]
- Kolodrubetz D., Schleif R. L-arabinose transport systems in Escherichia coli K-12. J Bacteriol. 1981 Nov;148(2):472–479. doi: 10.1128/jb.148.2.472-479.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolodrubetz D., Schleif R. Regulation of the L-arabinose transport operons in Escherichia coli. J Mol Biol. 1981 Sep 15;151(2):215–227. doi: 10.1016/0022-2836(81)90512-x. [DOI] [PubMed] [Google Scholar]
- Kunkel T. A., Roberts J. D., Zakour R. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- Lichenstein H. S., Hamilton E. P., Lee N. Repression and catabolite gene activation in the araBAD operon. J Bacteriol. 1987 Feb;169(2):811–822. doi: 10.1128/jb.169.2.811-822.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobell R. B., Schleif R. F. AraC-DNA looping: orientation and distance-dependent loop breaking by the cyclic AMP receptor protein. J Mol Biol. 1991 Mar 5;218(1):45–54. doi: 10.1016/0022-2836(91)90872-4. [DOI] [PubMed] [Google Scholar]
- Lobell R. B., Schleif R. F. DNA looping and unlooping by AraC protein. Science. 1990 Oct 26;250(4980):528–532. doi: 10.1126/science.2237403. [DOI] [PubMed] [Google Scholar]
- Martin K., Huo L., Schleif R. F. The DNA loop model for ara repression: AraC protein occupies the proposed loop sites in vivo and repression-negative mutations lie in these same sites. Proc Natl Acad Sci U S A. 1986 Jun;83(11):3654–3658. doi: 10.1073/pnas.83.11.3654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyle H., Waldburger C., Susskind M. M. Hierarchies of base pair preferences in the P22 ant promoter. J Bacteriol. 1991 Mar;173(6):1944–1950. doi: 10.1128/jb.173.6.1944-1950.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulligan M. E., McClure W. R. Analysis of the occurrence of promoter-sites in DNA. Nucleic Acids Res. 1986 Jan 10;14(1):109–126. doi: 10.1093/nar/14.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers R. M., Lerman L. S., Maniatis T. A general method for saturation mutagenesis of cloned DNA fragments. Science. 1985 Jul 19;229(4710):242–247. doi: 10.1126/science.2990046. [DOI] [PubMed] [Google Scholar]
- Ogden S., Haggerty D., Stoner C. M., Kolodrubetz D., Schleif R. The Escherichia coli L-arabinose operon: binding sites of the regulatory proteins and a mechanism of positive and negative regulation. Proc Natl Acad Sci U S A. 1980 Jun;77(6):3346–3350. doi: 10.1073/pnas.77.6.3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponnambalam S., Chan B., Busby S. Functional analysis of different sequence elements in the Escherichia coli galactose operon P2 promoter. Mol Microbiol. 1988 Mar;2(2):165–172. doi: 10.1111/j.1365-2958.1988.tb00018.x. [DOI] [PubMed] [Google Scholar]
- Ponnambalam S., Webster C., Bingham A., Busby S. Transcription initiation at the Escherichia coli galactose operon promoters in the absence of the normal -35 region sequences. J Biol Chem. 1986 Dec 5;261(34):16043–16048. [PubMed] [Google Scholar]
- Ren Y. L., Garges S., Adhya S., Krakow J. S. Cooperative DNA binding of heterologous proteins: evidence for contact between the cyclic AMP receptor protein and RNA polymerase. Proc Natl Acad Sci U S A. 1988 Jun;85(12):4138–4142. doi: 10.1073/pnas.85.12.4138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richet E., Vidal-Ingigliardi D., Raibaud O. A new mechanism for coactivation of transcription initiation: repositioning of an activator triggered by the binding of a second activator. Cell. 1991 Sep 20;66(6):1185–1195. doi: 10.1016/0092-8674(91)90041-v. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schleif R. An L-arabinose binding protein and arabinose permeation in Escherichia coli. J Mol Biol. 1969 Nov 28;46(1):185–196. doi: 10.1016/0022-2836(69)90065-5. [DOI] [PubMed] [Google Scholar]
- Schultz S. C., Shields G. C., Steitz T. A. Crystal structure of a CAP-DNA complex: the DNA is bent by 90 degrees. Science. 1991 Aug 30;253(5023):1001–1007. doi: 10.1126/science.1653449. [DOI] [PubMed] [Google Scholar]
- Scripture J. B., Voelker C., Miller S., O'Donnell R. T., Polgar L., Rade J., Horazdovsky B. F., Hogg R. W. High-affinity L-arabinose transport operon. Nucleotide sequence and analysis of gene products. J Mol Biol. 1987 Sep 5;197(1):37–46. doi: 10.1016/0022-2836(87)90607-3. [DOI] [PubMed] [Google Scholar]
- Shortle D., Nathans D. Local mutagenesis: a method for generating viral mutants with base substitutions in preselected regions of the viral genome. Proc Natl Acad Sci U S A. 1978 May;75(5):2170–2174. doi: 10.1073/pnas.75.5.2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegele D. A., Hu J. C., Walter W. A., Gross C. A. Altered promoter recognition by mutant forms of the sigma 70 subunit of Escherichia coli RNA polymerase. J Mol Biol. 1989 Apr 20;206(4):591–603. doi: 10.1016/0022-2836(89)90568-8. [DOI] [PubMed] [Google Scholar]
- Stoltzfus L., Wilcox G. Effect of mutations in the cyclic AMP receptor protein-binding site on araBAD and araC expression. J Bacteriol. 1989 Feb;171(2):1178–1184. doi: 10.1128/jb.171.2.1178-1184.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoner C., Schleif R. The araE low affinity L-arabinose transport promoter. Cloning, sequence, transcription start site and DNA binding sites of regulatory proteins. J Mol Biol. 1983 Dec 25;171(4):369–381. doi: 10.1016/0022-2836(83)90035-9. [DOI] [PubMed] [Google Scholar]
- Tomizawa J. Control of ColE1 plasmid replication: initial interaction of RNA I and the primer transcript is reversible. Cell. 1985 Mar;40(3):527–535. doi: 10.1016/0092-8674(85)90201-6. [DOI] [PubMed] [Google Scholar]
- Valentin-Hansen P., Holst B., Søgaard-Andersen L., Martinussen J., Nesvera J., Douthwaite S. R. Design of cAMP-CRP-activated promoters in Escherichia coli. Mol Microbiol. 1991 Feb;5(2):433–437. doi: 10.1111/j.1365-2958.1991.tb02126.x. [DOI] [PubMed] [Google Scholar]
- Waldburger C., Gardella T., Wong R., Susskind M. M. Changes in conserved region 2 of Escherichia coli sigma 70 affecting promoter recognition. J Mol Biol. 1990 Sep 20;215(2):267–276. doi: 10.1016/s0022-2836(05)80345-6. [DOI] [PubMed] [Google Scholar]
- Wilcox G., Meuris P., Bass R., Englesberg E. Regulation of the L-arabinose operon BAD in vitro. J Biol Chem. 1974 May 10;249(9):2946–2952. [PubMed] [Google Scholar]
- Youderian P., Bouvier S., Susskind M. M. Sequence determinants of promoter activity. Cell. 1982 Oct;30(3):843–853. doi: 10.1016/0092-8674(82)90289-6. [DOI] [PubMed] [Google Scholar]
- Zhang X. P., Ebright R. H. Identification of a contact between arginine-180 of the catabolite gene activator protein (CAP) and base pair 5 of the DNA site in the CAP-DNA complex. Proc Natl Acad Sci U S A. 1990 Jun;87(12):4717–4721. doi: 10.1073/pnas.87.12.4717. [DOI] [PMC free article] [PubMed] [Google Scholar]