Abstract
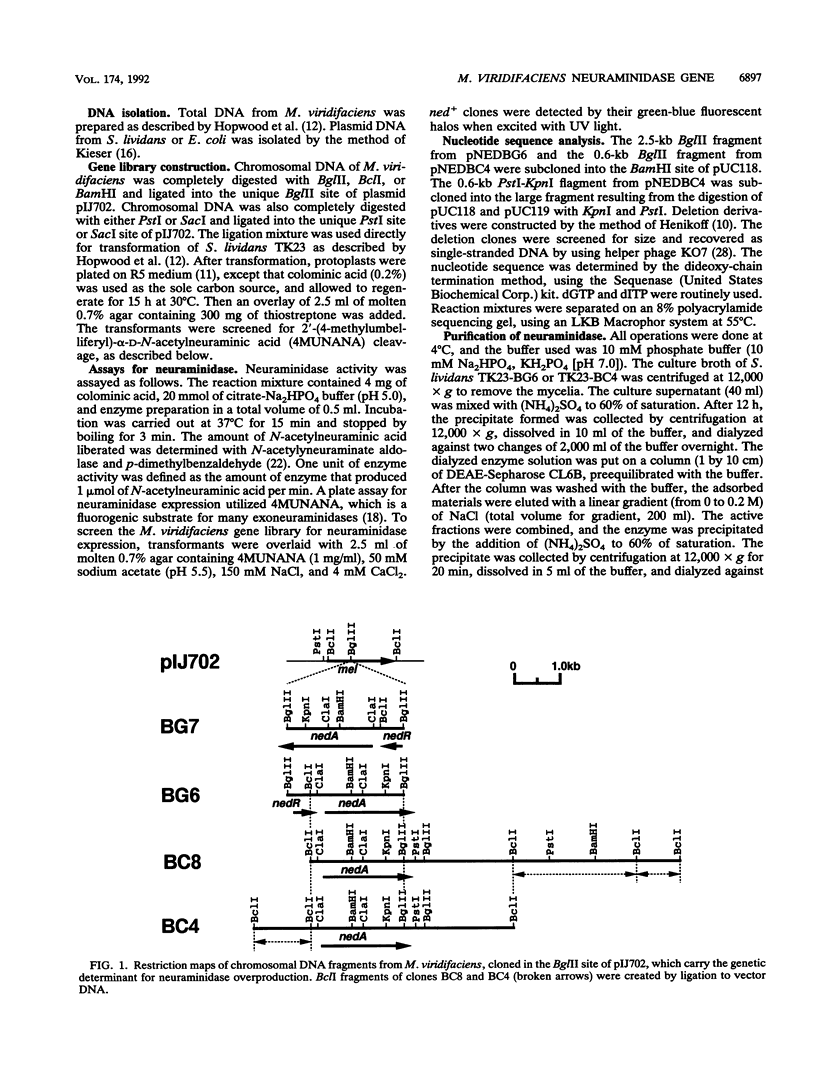
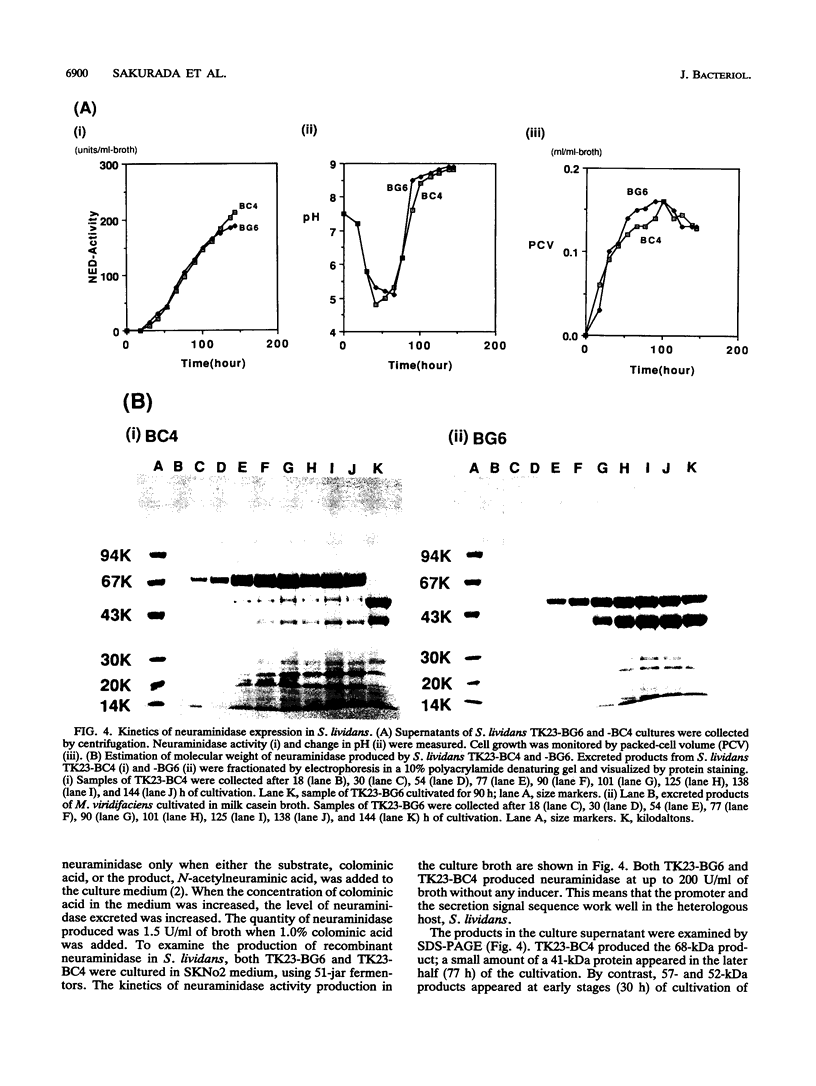
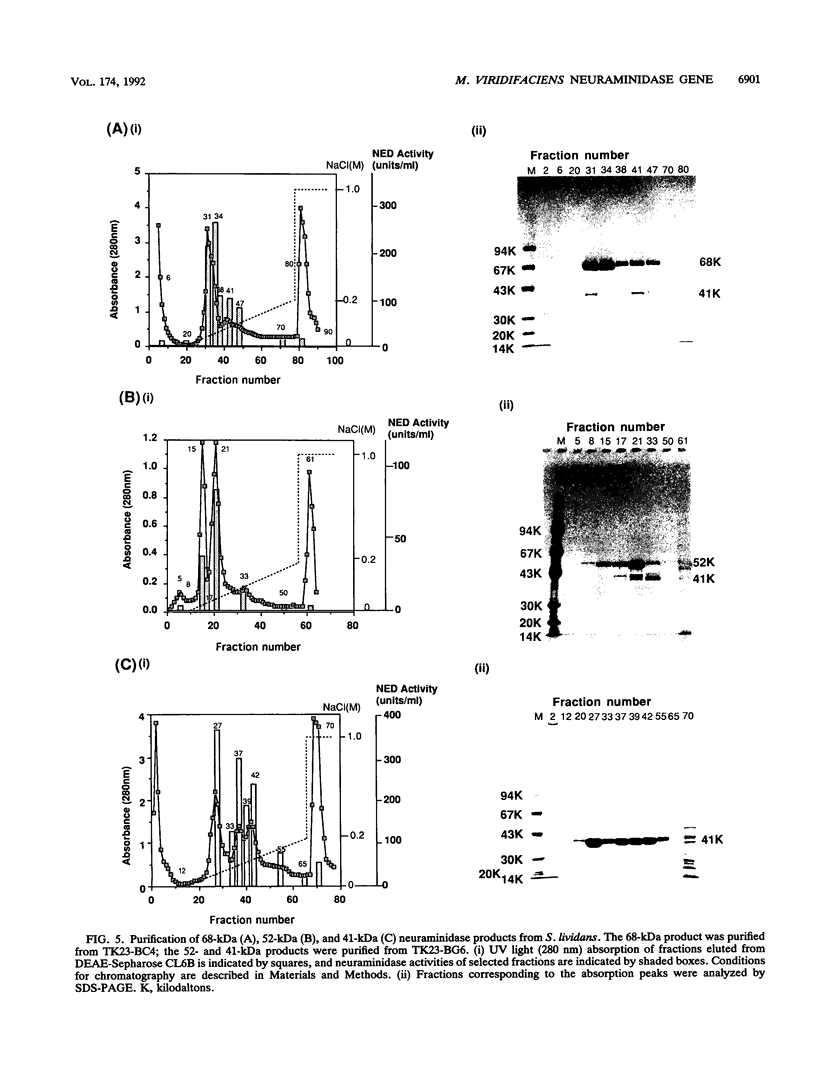
We have cloned the Micromonospora viridifaciens neuraminidase (EC 3.2.1.18) gene (nedA) in Streptomyces lividans. This was accomplished by using the vector pIJ702 and BglII-BclI libraries of M. viridifaciens chromosomal inserts created in S. lividans. The libraries were screened for the expression of neuraminidase by monitoring the cleavage of the fluorogenic neuraminidase substrate 2'-(4-methylumbelliferyl)-alpha-D-N-acetyl-neuraminic acid. Positive clones (BG6, BG7, BC4, and BC8) contained the identical 2-kb BclI-BglII fragment and expressed neuraminidase efficiently and constitutively using its own promoter in the heterologous host. From the nucleotide sequence analysis, an open reading frame of 1,941 bp which encodes a polypeptide with an M(r) of 68,840 was detected. The deduced amino acid sequence has five Asp boxes, -Ser-X-Asp-X-Gly-X-Thr-Trp, showing great similarity to other bacterial and viral neuraminidases. We have also identified the catalytic domain by using truncated proteins produced in S. lividans.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Air G. M., Laver W. G. The neuraminidase of influenza virus. Proteins. 1989;6(4):341–356. doi: 10.1002/prot.340060402. [DOI] [PubMed] [Google Scholar]
- Bibb M. J., Findlay P. R., Johnson M. W. The relationship between base composition and codon usage in bacterial genes and its use for the simple and reliable identification of protein-coding sequences. Gene. 1984 Oct;30(1-3):157–166. doi: 10.1016/0378-1119(84)90116-1. [DOI] [PubMed] [Google Scholar]
- Bibb M. J., Janssen G. R., Ward J. M. Cloning and analysis of the promoter region of the erythromycin resistance gene (ermE) of Streptomyces erythraeus. Gene. 1985;38(1-3):215–226. doi: 10.1016/0378-1119(85)90220-3. [DOI] [PubMed] [Google Scholar]
- Fickett J. W. Recognition of protein coding regions in DNA sequences. Nucleic Acids Res. 1982 Sep 11;10(17):5303–5318. doi: 10.1093/nar/10.17.5303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison S. C., Aggarwal A. K. DNA recognition by proteins with the helix-turn-helix motif. Annu Rev Biochem. 1990;59:933–969. doi: 10.1146/annurev.bi.59.070190.004441. [DOI] [PubMed] [Google Scholar]
- Hasegawa M., Dairi T., Ohta T., Hashimoto E. A novel, highly efficient gene-cloning system for Micromonospora strains. J Bacteriol. 1991 Nov;173(21):7004–7011. doi: 10.1128/jb.173.21.7004-7011.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heffron F., McCarthy B. J., Ohtsubo H., Ohtsubo E. DNA sequence analysis of the transposon Tn3: three genes and three sites involved in transposition of Tn3. Cell. 1979 Dec;18(4):1153–1163. doi: 10.1016/0092-8674(79)90228-9. [DOI] [PubMed] [Google Scholar]
- Henikoff S. Unidirectional digestion with exonuclease III creates targeted breakpoints for DNA sequencing. Gene. 1984 Jun;28(3):351–359. doi: 10.1016/0378-1119(84)90153-7. [DOI] [PubMed] [Google Scholar]
- Hintermann G., Crameri R., Vögtli M., Hütter R. Streptomycin-sensitivity in Streptomyces glaucescens is due to deletions comprising the structural gene coding for a specific phosphotransferase. Mol Gen Genet. 1984;196(3):513–520. doi: 10.1007/BF00436201. [DOI] [PubMed] [Google Scholar]
- Hopwood D. A., Bibb M. J., Chater K. F., Kieser T. Plasmid and phage vectors for gene cloning and analysis in Streptomyces. Methods Enzymol. 1987;153:116–166. doi: 10.1016/0076-6879(87)53052-x. [DOI] [PubMed] [Google Scholar]
- Hoshiko S., Makabe O., Nojiri C., Katsumata K., Satoh E., Nagaoka K. Molecular cloning and characterization of the Streptomyces hygroscopicus alpha-amylase gene. J Bacteriol. 1987 Mar;169(3):1029–1036. doi: 10.1128/jb.169.3.1029-1036.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz E., Thompson C. J., Hopwood D. A. Cloning and expression of the tyrosinase gene from Streptomyces antibioticus in Streptomyces lividans. J Gen Microbiol. 1983 Sep;129(9):2703–2714. doi: 10.1099/00221287-129-9-2703. [DOI] [PubMed] [Google Scholar]
- Kieser T. Factors affecting the isolation of CCC DNA from Streptomyces lividans and Escherichia coli. Plasmid. 1984 Jul;12(1):19–36. doi: 10.1016/0147-619x(84)90063-5. [DOI] [PubMed] [Google Scholar]
- King J., Laemmli U. K. Polypeptides of the tail fibres of bacteriophage T4. J Mol Biol. 1971 Dec 28;62(3):465–477. doi: 10.1016/0022-2836(71)90148-3. [DOI] [PubMed] [Google Scholar]
- Myers R. W., Lee R. T., Lee Y. C., Thomas G. H., Reynolds L. W., Uchida Y. The synthesis of 4-methylumbelliferyl alpha-ketoside of N-acetylneuraminic acid and its use in a fluorometric assay for neuraminidase. Anal Biochem. 1980 Jan 1;101(1):166–174. doi: 10.1016/0003-2697(80)90056-1. [DOI] [PubMed] [Google Scholar]
- Nakai R., Horinouchi S., Beppu T. Cloning and nucleotide sequence of a cellulase gene, casA, from an alkalophilic Streptomyces strain. Gene. 1988 May 30;65(2):229–238. doi: 10.1016/0378-1119(88)90459-3. [DOI] [PubMed] [Google Scholar]
- Nees S., Veh R. W., Schauer R. Purification and characterization of neuraminidase from Clostridium perfringens. Hoppe Seylers Z Physiol Chem. 1975 Jun;356(6):1027–1042. doi: 10.1515/bchm2.1975.356.s1.1027. [DOI] [PubMed] [Google Scholar]
- Pabo C. O., Sauer R. T. Protein-DNA recognition. Annu Rev Biochem. 1984;53:293–321. doi: 10.1146/annurev.bi.53.070184.001453. [DOI] [PubMed] [Google Scholar]
- REISSIG J. L., STORMINGER J. L., LELOIR L. F. A modified colorimetric method for the estimation of N-acetylamino sugars. J Biol Chem. 1955 Dec;217(2):959–966. [PubMed] [Google Scholar]
- Roggentin P., Rothe B., Kaper J. B., Galen J., Lawrisuk L., Vimr E. R., Schauer R. Conserved sequences in bacterial and viral sialidases. Glycoconj J. 1989;6(3):349–353. doi: 10.1007/BF01047853. [DOI] [PubMed] [Google Scholar]
- Roggentin P., Rothe B., Lottspeich F., Schauer R. Cloning and sequencing of a Clostridium perfringens sialidase gene. FEBS Lett. 1988 Sep 26;238(1):31–34. doi: 10.1016/0014-5793(88)80219-9. [DOI] [PubMed] [Google Scholar]
- Rothe B., Roggentin P., Frank R., Blöcker H., Schauer R. Cloning, sequencing and expression of a sialidase gene from Clostridium sordellii G12. J Gen Microbiol. 1989 Nov;135(11):3087–3096. doi: 10.1099/00221287-135-11-3087. [DOI] [PubMed] [Google Scholar]
- Russo T. A., Thompson J. S., Godoy V. G., Malamy M. H. Cloning and expression of the Bacteroides fragilis TAL2480 neuraminidase gene, nanH, in Escherichia coli. J Bacteriol. 1990 May;172(5):2594–2600. doi: 10.1128/jb.172.5.2594-2600.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira J., Messing J. Production of single-stranded plasmid DNA. Methods Enzymol. 1987;153:3–11. doi: 10.1016/0076-6879(87)53044-0. [DOI] [PubMed] [Google Scholar]
- Vimr E. R., Lawrisuk L., Galen J., Kaper J. B. Cloning and expression of the Vibrio cholerae neuraminidase gene nanH in Escherichia coli. J Bacteriol. 1988 Apr;170(4):1495–1504. doi: 10.1128/jb.170.4.1495-1504.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vimr E. R., Troy F. A. Identification of an inducible catabolic system for sialic acids (nan) in Escherichia coli. J Bacteriol. 1985 Nov;164(2):845–853. doi: 10.1128/jb.164.2.845-853.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]